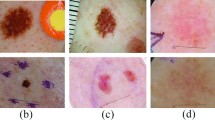
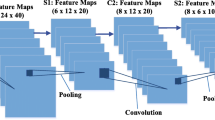
Abstract
The automatic segmentation of skin lesions in dermoscopic images is a challenging task due to the presence of artifacts, small lesion sizes, and low contrast between lesions and non-lesion regions. Deep learning models such as U-Net have been used for accurate segmentation in general. Still, their success rate is limited in the case of dermoscopic images due to these challenges. In this paper, we propose a U-Net-based model for efficient and effective segmentation of skin lesions in dermoscopic images. The proposed model, called Attention Residual U-Net with a modified decoder (ARU-Net-MD), employs an encoder-decoder architecture with residual learning, attention gates, and a modified decoder with a combined loss function to achieve higher accuracy for the semantic segmentation of dermoscopic images. Residual learning allows for an efficient model with fewer parameters, while attention gates highlight important features, and the modified decoder with a combined loss function assists the learning process and enables the model to learn more semantic information during training. We evaluated our model on four publicly available datasets, PH2, ISIC 2016, ISIC 2017, and ISIC 2018, and observed an accuracy of 0.96, 0.97, 0.95, and 0.96, respectively, outperforming other state-of-the-art skin lesion segmentation models.









Similar content being viewed by others
Data availability
The datasets analyzed during the current study are PH2, ISIC 2016, and ISIC 2018. PH2 dataset is publicly available at https://www.fc.up.pt/addi/ph2%20database.html, and ISIC 2016, ISIC 2017, and ISIC 2018 are available at https://challenge.isic-archive.com/data/.
References
Iqbal I, Younus M, Walayat K, Kakar MU, Ma J (2021) Automated multi-class classification of skin lesions through deep convolutional neural network with dermoscopic images. Comput Med Imaging Graph 88:101843. https://doi.org/10.1016/j.compmedimag.2020.101843
Siegel RL, Miller KD, Wagle NS, Jemal A (2023) Cancer statistics, 2023. Ca Cancer J Clin 73(1):17–48. https://doi.org/10.3322/caac.21763
Stenzel AE, Miller J, Holtan SG, Brown K, Ahmed RL, Lazovich D, Vogel RI (2023) Cross-sectional study of physical activity among long-term melanoma survivors and population controls. Arch Dermatol Res 315(4):1011–1016. https://doi.org/10.1007/s00403-022-02334-2
Nachbar F, Stolz W, Merkle T, Cognetta AB, Vogt T, Landthaler M, Bilek P, Braun-Falco O, Plewig G (1994) The ABCD rule of dermatoscopy: high prospective value in the diagnosis of doubtful melanocytic skin lesions. J Am Acad Dermatol 30(4):551–559. https://doi.org/10.1016/S0190-9622(94)70061-3
Garnavi R, Aldeen M, Celebi ME, Bhuiyan A, Dolianitis C, Varigos G (2009) Skin lesion segmentation using color channel optimization and clustering-based histogram thresholding. Int J Biomed Biol Eng 3(12):365–373. https://doi.org/10.5281/zenodo.1072764
Schaefer G, Rajab MI, Celebi ME, Iyatomi H (2011) Colour and contrast enhancement for improved skin lesion segmentation. Comput Med Imaging Graph 35(2):99–104. https://doi.org/10.1016/j.compmedimag.2010.08.004
Celebi ME, Kingravi HA, Uddin B, Iyatomi H, Aslandogan YA, Stoecker WV, Moss RH (2007) A methodological approach to the classification of dermoscopy images. Comput Med Imaging Graph 31(6):362–373. https://doi.org/10.1016/j.compmedimag.2007.01.003
Wong A, Scharcanski J, Fieguth P (2011) Automatic skin lesion segmentation via iterative stochastic region merging. IEEE Trans Inf Technol Biomed 15(6):929–936. https://doi.org/10.1109/TITB.2011.2157829
Abbas Q, Fondón I, Rashid M (2011) Unsupervised skin lesions border detection via two-dimensional image analysis. Comput Methods Programs Biomed 104(3):1–5. https://doi.org/10.1016/j.cmpb.2010.06.016
Sforza G, Castellano G, Arika SK, LeAnder RW, Stanley RJ, Stoecker WV, Hagerty JR (2012) Using adaptive thresholding and skewness correction to detect gray areas in melanoma in situ images. IEEE Trans Instrum Meas 61(7):1839–1847. https://doi.org/10.1109/TIM.2012.2192349
Castillejos H, Ponomaryov V, Nino-de-Rivera L, Golikov V (2012) Wavelet transform fuzzy algorithms for dermoscopic image segmentation. Comput Math Methods Med 2012:1. https://doi.org/10.1155/2012/578721
Zhou H, Schaefer G, Sadka AH, Celebi ME (2009) Anisotropic mean shift based fuzzy c-means segmentation of dermoscopy images. IEEE J Selec Topics Signal Process 3(1):26–34. https://doi.org/10.1109/JSTSP.2008.2010631
Jaisakthi SM, Mirunalini P, Aravindan C (2018) Automated skin lesion segmentation of dermoscopic images using GrabCut and k-means algorithms. IET Comput Vis (8):1088–1095. https://doi.org/10.1049/iet-cvi.2018.5289
Oliveira RB, Mercedes Filho E, Ma Z, Papa JP, Pereira AS, Tavares JM (2016) Computational methods for the image segmentation of pigmented skin lesions: a review. Comput Methods Programs Biomed 131:127–141. https://doi.org/10.1016/j.cmpb.2016.03.032
ul haq I, Amin J, Sharif M, Almas Anjum M (2022) Skin lesion detection using recent machine learning approaches. In: Prognostic models in healthcare: AI and Statistical approaches. Springer Nature Singapore, Singapore, pp 193–211. https://doi.org/10.1007/978-981-19-2057-8_7
Murugan A, Nair SA, Kumar KS (2019) Detection of skin cancer using SVM, random forest and kNN classifiers. J Med Syst 43:1–9. https://doi.org/10.1007/s10916-019-1400-8
Taufiq MA, Hameed N, Anjum A, Hameed F (2016) m-Skin doctor: a mobile enabled system for early melanoma skin cancer detection using support vector machine. eHealth 360°: International Summit on eHealth, Budapest, Hungary. Springer International Publishing, pp 468–475. https://doi.org/10.1007/978-3-319-49655-9_57
Akram T, Khan MA, Sharif M, Yasmin M (2018) Skin lesion segmentation and recognition using multichannel saliency estimation and M-SVM on selected serially fused features. J Ambient Intell Humaniz Comput: 1–20. https://doi.org/10.1007/s12652-018-1051-5
Nazi ZA, Abir TA (2020) Automatic skin lesion segmentation and melanoma detection: transfer learning approach with u-net and dcnn-svm. In: Proceedings of International Joint Conference on Computational Intelligence: IJCCI 2018. Springer, Singapore, pp 371–381. https://doi.org/10.1007/978-981-13-7564-4_32
Bassel A, Abdulkareem AB, Alyasseri ZA, Sani NS, Mohammed HJ (2022) Automatic malignant and benign skin cancer classification using a hybrid deep learning approach. Diagnostics 12(10):2472. https://doi.org/10.3390/diagnostics12102472
Monika MK, Vignesh NA, Kumari CU, Kumar MN, Lydia EL (2020) Skin cancer detection and classification using machine learning. Mater Today: Proc 33:4266–4270. https://doi.org/10.1016/j.matpr.2020.07.366
Kayalibay B, Jensen G, van der Smagt P (2017) CNN-based segmentation of medical imaging data. ar**v preprint ar**v:1701.03056. https://doi.org/10.48550/ar**v.1701.03056
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. InMedical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, Proceedings, Part III, Springer International Publishing, pp 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Zhang Z, Liu Q, Wang Y (2018) Road extraction by deep residual u-net. IEEE Geosci Remote Sens Lett 15(5):749–753. https://doi.org/10.1109/LGRS.2018.2802944
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, Glocker B (2018) Attention u-net: learning where to look for the pancreas. ar**v Preprint ar**v 180403999. https://doi.org/10.48550/ar**v.1804.03999
Schuster M, Paliwal KK (1997) Bidirectional recurrent neural networks. IEEE Trans Signal Process 45(11):2673–2681. https://doi.org/10.1109/78.650093
Girshick R, Donahue J, Darrell T, Malik J (2015) Region-based convolutional networks for accurate object detection and segmentation. IEEE Trans Pattern Anal Mach Intell 38(1):142–158. https://doi.org/10.1109/TPAMI.2015.2437384
Alom MZ, Hasan M, Yakopcic C, Taha TM, Asari VK (2018) Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. ar**. ar**v preprint ar**v:2305.11003. https://doi.org/10.48550/ar**v.2305.11003
Ni ZL, Bian GB, Zhou XH, Hou ZG, **e XL, Wang C, Zhou YJ, Li RQ, Li Z (2019) Raunet: Residual attention u-net for semantic segmentation of cataract surgical instruments. InInternational Conference on Neural Information Processing. Springer International Publishing, Cham, pp 139–149. https://doi.org/10.1007/978-3-030-36711-4_13
Chen LC, Yang Y, Wang J, Xu W, Yuille AL (2016) Attention to scale: scale-aware semantic image segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 3640–3649. https://doi.org/10.1109/CVPR.2016.396
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Yu L, Chen H, Dou Q, Qin J, Heng PA (2016) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging 36(4):994–1004. https://doi.org/10.1109/TMI.2016.2642839
Yuan Y, Chao M, Lo YC (2017) Automatic skin lesion segmentation using deep fully convolutional networks with jaccard distance. IEEE Trans Med Imaging 36(9):1876–1886. https://doi.org/10.1109/TMI.2017.2695227
Yuan Y, Lo YC (2017) Improving dermoscopic image segmentation with enhanced convolutional-deconvolutional networks. IEEE J Biomed Health Inf 23(2):519–526. https://doi.org/10.1109/JBHI.2017.2787487
Venkatesh GM, Naresh YG, Little S, O’Connor NE (2018) A deep residual architecture for skin lesion segmentation. InOR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis: First International Workshop, OR 2.0 2018, 5th International Workshop, CARE 2018, 7th International Workshop, CLIP 2018, Third International Workshop, ISIC 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, Springer International Publishing, pp 277–284. https://doi.org/10.1007/978-3-030-01201-4_30
Dash M, Londhe ND, Ghosh S, Semwal A, Sonawane RS (2019) PsLSNet: automated psoriasis skin lesion segmentation using modified u-net-based fully convolutional network. Biomed Signal Process Control 52:226–237. https://doi.org/10.1016/j.bspc.2019.04.002
Tang P, Liang Q, Yan X, **ang S, Sun W, Zhang D, Coppola G (2019) Efficient skin lesion segmentation using separable-unet with stochastic weight averaging. Comput Methods Programs Biomed 178:289–301. https://doi.org/10.1016/j.cmpb.2019.07.005
Goyal M, Oakley A, Bansal P, Dancey D, Yap MH (2019) Skin lesion segmentation in dermoscopic images with ensemble deep learning methods. IEEE Access 8:4171–4181. https://doi.org/10.1109/ACCESS.2019.2960504
Lin D, Li Y, Nwe TL, Dong S, Oo ZM (2020) RefineU-Net: Improved U-Net with progressive global feedbacks and residual attention guided local refinement for medical image segmentation. Pattern Recognit Lett 138:267–275. https://doi.org/10.1016/j.patrec.2020.07.013
Arora R, Raman B, Nayyar K, Awasthi R (2021) Automated skin lesion segmentation using attention-based deep convolutional neural network. Biomed Signal Process Control 65:102358. https://doi.org/10.1016/j.bspc.2020.102358
Wibowo A, Purnama SR, Wirawan PW, Rasyidi H (2021) Lightweight encoder-decoder model for automatic skin lesion segmentation. Inf Med Unlocked 25:100640. https://doi.org/10.1016/j.imu.2021.100640
Sarker MM, Rashwan HA, Akram F, Singh VK, Banu SF, Chowdhury FU, Choudhury KA, Chambon S, Radeva P, Puig D, Abdel-Nasser M (2021) SLSNet: skin lesion segmentation using a lightweight generative adversarial network. Expert Syst Appl 183:115433. https://doi.org/10.1016/j.eswa.2021.115433
Tong X, Wei J, Sun B, Su S, Zuo Z, Wu P (2021) ASCU-Net: attention gate, spatial and channel attention u-net for skin lesion segmentation. Diagnostics 11(3):501. https://doi.org/10.3390/diagnostics11030501
Wang Y, Wang S (2022) Skin lesion segmentation with attention-based SC-Conv U-Net and feature map distortion. SIViP 6:1471–1479. https://doi.org/10.1007/s11760-021-02100-3
Khouloud S, Ahlem M, Fadel T, Amel S (2022) W-net and inception residual network for skin lesion segmentation and classification. Appl Intell 1–9.https://doi.org/10.1007/s10489-021-02652-4
Wu H, Pan J, Li Z, Wen Z, Qin J (2020) Automated skin lesion segmentation via an adaptive dual attention module. IEEE Trans Med Imaging 40(1):357–370. https://doi.org/10.1109/TMI.2020.3027341
Qiu S, Li C, Feng Y, Zuo S, Liang H, Xu A (2023) GFANet: gated fusion attention network for skin lesion segmentation. Comput Biol Med 155:106462. https://doi.org/10.1016/j.compbiomed.2022.106462
Zeng G, Peng H, Li A, Liu Z, Liu C, Yu PS, He L (2023) Unsupervised skin lesion segmentation via structural entropy minimization on multi-scale superpixel graphs. ar**v preprint ar**v:2309.01899. https://doi.org/10.48550/ar**v.2309.01899
Li X, Peng B, Hu J, Ma C, Yang D, **e Z (2024) USL-Net: uncertainty self-learning network for unsupervised skin lesion segmentation. Biomed Signal Process Control 89:105769. https://doi.org/10.1016/j.bspc.2023.105769
Agarap AF (2018) Deep learning using rectified linear units (relu). ar**v preprint ar**v:1803.08375. https://doi.org/10.48550/ar**v.1803.08375
Rajon DA, Bolch WE (2003) Marching cube algorithm: review and trilinear interpolation adaptation for image-based dosimetric models. Comput Med Imaging Graph 27(5):411–435. https://doi.org/10.1016/S0895-6111(03)00032-6
Lin TY, Goyal P, Girshick R, He K, Dollár P (2018) Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell 42(2):318–327. https://doi.org/10.1109/TPAMI.2018.2858826
Jadon S (2020) A survey of loss functions for semantic segmentation. In: 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), pp 1–7. https://doi.org/10.1109/CIBCB48159.2020.9277638
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. ar**v preprint ar**v:1412.6980. https://doi.org/10.48550/ar**v.1412.6980
Mendonça T, Ferreira PM, Marques JS, Marcal AR, Rozeira J (2013) PH 2-A dermoscopic image database for research and benchmarking. In: 35th annual international conference of the IEEE engineering in medicine and biology society (EMBC), pp 5437–5440. https://doi.org/10.1109/EMBC.2013.6610779
Gutman D, Codella NC, Celebi E, Helba B, Marchetti M, Mishra N, Halpern A (2016) Skin lesion analysis toward melanoma detection: A challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC). ar**v preprint ar**v:1605.01397. https://doi.org/10.48550/ar**v.1605.01397
Codella NC, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H, Halpern A (2018) Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018) pp 168–172. https://doi.org/10.1109/ISBI.2018.8363547
Codella N, Rotemberg V, Tschandl P, Celebi ME, Dusza S, Gutman D, Helba B, Kalloo A, Liopyris K, Marchetti M, Kittler H (2019) Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). ar**v preprint ar**v:1902.03368. https://doi.org/10.48550/ar**v.1902.03368
He C, Li K, Xu G, Yan J, Tang L, Zhang Y, Wang Y, Li X (2023) Hqg-net: unpaired medical image enhancement with high-quality guidance. IEEE Trans Neural Netw Learn Syst. https://doi.org/10.1109/TNNLS.2023.3315307
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All the authors contributed equally to this work.
Corresponding author
Ethics declarations
Ethics approval
The research does not involve any human or animal participant, their data, or biological material. No ethical approval is required.
Consent to participate
The research does not involve any human individual participant. Consent to participate is not required.
Consent for publication
The manuscript does not contain any individual person’s data in any form. Consent to publish is not required.
Conflict of interest/Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kaur, R., Kaur, S. Automatic skin lesion segmentation using attention residual U-Net with improved encoder-decoder architecture. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18895-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18895-5