Abstract
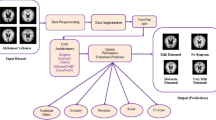
Alzheimer’s disease (AD) is a neurodegenerative disease that is well-known for causing continuous loss of memory, cognition, and other higher brain functions. AD is not a single disease, but rather a group of related diseases with similar characteristics. The use of deep neural network-based pattern classification techniques, such as convolutional neural networks, is effective in classifying patients into different sub-types of AD and in distinguishing the different stages of severity of the disease. in the medical field, early detection of its start can be quite beneficial. This article focuses on the early detection of various stages of cognitive aging and AD using neuroimaging and transfer learning (TL). Images of imagery via resonance magnetic (IRM) obtained from a Kaggle database called Alzheimer’s Dataset ( 4 class of Images) with several classes of non-dementia (NONDEM), very mild dementia(VERDEM), mild dementia(MILDEM), moderate dementia(MODDEM) are classified using a transfer learning approach. In this work, we compare the classification performance of six pre-trained networks, which are VGG-19, VGG-16, ResNet-50, InceptionV3, Xception, and DenseNet169. They were enthralled and tested using 6400 images from the Kaggle data pool. The confusion matrix and its parameters are used to assess the classification performance of these six networks. VGG-19, VGG-16, Inception-V3, Xception, ResNet-50, and DenseNet169 all have 92.86%, 92.83%, 91.04%, 90.57%, 85.99%, and 88.64% overall precision in MA detection, respectively.














Similar content being viewed by others
Data availability statement
Data supporting the findings of this study are freely available in [Kaggle] at [https://www.kaggle.com/datasets/tourist55/alzheimers-dataset-4-class-of-images], reference number [40]
References
Sudharsan M, Thailambal G (2021) Alzheimer’s disease prediction using machine learning techniques and principal component analysis (pca). Materials today: proceedings
O’Shea K, Nash R (2015) An introduction to convolutional neural networks. ar**v preprint ar**v:1511.08458
Aderghal K, Khvostikov A, Krylov A, Benois-Pineau J, Afdel K, Catheline G (2018) Classification of alzheimer disease on imaging modalities with deep cnns using cross-modal transfer learning. In: 2018 IEEE 31st International symposium on computer-based medical systems (CBMS). IEEE, pp 345–350
Buvaneswari P, Gayathri R (2021) Deep learning-based segmentation in classification of alzheimer’s disease. Arab J Sci Eng 46(6):5373–5383
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39(12):2481–2495
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Al-Adhaileh MH (2022) Diagnosis and classification of alzheimer’s disease by using a convolution neural network algorithm. Soft Computing, 1–12
Krizhevsky A, Sutskever I, Hinton GE (2017) Imagenet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
Sambath Kumar S, Nandhini M (2022) Automated classification of alzheimer’s disease using mri and transfer learning. In: Mobile computing and sustainable informatics. Springer, ???, pp 663–686
Kumari A, Tanwar S, Tyagi S, Kumar N (2018) Fog computing for healthcare 4.0 environment: opportunities and challenges. Computers & Electrical Engineering 72:1–13
Tanwar S, Kumari A, Vekaria D, Kumar N, Sharma R (2022) An ai-based disease detection and prevention scheme for covid-19. Comput Electr Eng 103:108352
Basaia S, Agosta F, Wagner L, Canu E, Magnani G, Santangelo R, Filippi M, Initiative ADN et al (2019) Automated classification of alzheimer’s disease and mild cognitive impairment using a single mri and deep neural networks. NeuroImage: Clinical 21:101645
Liu M, Li F, Yan H, Wang K, Ma Y, Shen L, Xu M, Initiative ADN et al (2020) A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in alzheimer’s disease. Neuroimage 208:116459
Bui TD, Shin J, Moon T (2017) 3d densely convolutional networks for volumetric segmentation. ar**v preprint ar**v:1709.03199
Petersen RC, Aisen P, Beckett LA, Donohue M, Gamst A, Harvey DJ, Jack C, Jagust W, Shaw L, Toga A et al (2010) Alzheimer’s disease neuroimaging initiative (adni): clinical characterization. Neurology 74(3):201–209
Alinsaif S, Lang J, Initiative ADN et al (2021) 3d shearlet-based descriptors combined with deep features for the classification of alzheimer’s disease based on mri data. Comput Biol Med 138:104879
Patel VM, Easley GR, Healy DM (2009) Shearlet-based deconvolution. IEEE Trans Image Process 18(12):2673–2685
Wu H, Luo J, Lu X, Zeng Y (2022) 3d transfer learning network for classification of alzheimer’s disease with mri. International Journal of Machine Learning and Cybernetics, 1–15
Shanmugam JV, Duraisamy B, Simon BC, Bhaskaran P (2022) Alzheimer’s disease classification using pre-trained deep networks. Biomed Signal Process Control 71:103217
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL (2007) Open access series of imaging studies (oasis): cross-sectional mri data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci 19(9):1498–1507
Haque IRI, Neubert J (2020) Deep learning approaches to biomedical image segmentation. Informatics in Medicine Unlocked 18:100297
Wu J (2017) Introduction to convolutional neural networks. National Key Lab for Novel Software Technology. Nan**g University. China 5(23):495
Wu H, Gu X (2015) Max-pooling dropout for regularization of convolutional neural networks. In: International conference on neural information processing. Springer, pp 46–54
Sainath TN, Vinyals O, Senior A, Sak H (2015) Convolutional, long short-term memory, fully connected deep neural networks. In: 2015 IEEE International conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 4580–4584
Ying X (2019) An overview of overfitting and its solutions. In: Journal of physics: conference series, vol 1168. IOP Publishing, p 022022
Agarap AF (2018) Deep learning using rectified linear units (relu). ar**v preprint ar**v:1803.08375
Banerjee K, Gupta RR, Vyas K, Mishra B et al (2020) Exploring alternatives to softmax function. ar**v preprint ar**v:2011.11538
Tan C, Sun F, Kong T, Zhang W, Yang C, Liu C (2018) A survey on deep transfer learning. In: International conference on artificial neural networks. Springer, pp 270–279
Zhuang F, Qi Z, Duan K, ** D, Zhu Y, Zhu H, **ong H, He Q (2021) A comprehensive survey on transfer learning. Proc IEEE 109(1):43–76. https://doi.org/10.1109/JPROC.2020.3004555
Subetha T, Khilar R, Christo MS (2021) A comparative analysis on plant pathology classification using deep learning architecture–resnet and vgg19. Materials today: proceedings
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Varshni D, Thakral K, Agarwal L, Nijhawan R, Mittal A (2019) Pneumonia detection using cnn based feature extraction. In: 2019 IEEE International conference on electrical, computer and communication technologies (ICECCT). IEEE, pp 1–7
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1251–1258
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. ar**v preprint ar**v:1409.1556
Rai HM, Chatterjee K (2020) Detection of brain abnormality by a novel lu-net deep neural cnn model from mr images. Machine Learning with Applications 2:100004
Gandhi ST (2020) Context Sensitive Image Denoising and Enhancement Using U-Nets. Rochester Institute of Technology, ???
Lipton ZC, Elkan C, Narayanaswamy B (2014) Thresholding classifiers to maximize f1 score. ar**v preprint ar**v:1402.1892
Sarvesh dubey, (2019, december). alzheimer’s dataset (4 class of images) retrived march 2022 from https://www.kaggle.com/datasets/tourist55/alzheimers-dataset-4-class-of-images
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. ar**v preprint ar**v:1412.6980
Acknowledgements
The authors would like to thank the National Center for Scientific and Technical Research (CNRST) for supporting and funding this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest statement
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Assmi, A., Elhabyb, K., Benba, A. et al. Alzheimer’s disease classification: a comprehensive study. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18306-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18306-9