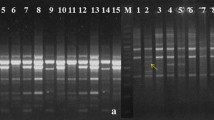
Abstract
Background
Embelia ribes Burm f. (Primulaceae) is a medicinal and vulnerable woody liana distributed throughout India. Embelin, a well-recognized active phytoconstituents in berries, is commonly used in ayurvedic formulations. Due to over-exploitation, the status of the plant is vulnerable. Previous studies on this species mainly focused on its phytochemical analysis, which led to overexploitation and loss of the germplasm.
Methods and results
In the present study, 20 RAPD and 18 ISSR markers were employed to assess genetic divergence in 40 genotypes of E. ribes collected from different parts of the Western Ghats of India. In RAPD analysis, all 40 accessions with 20 RAPD primers amplified 282 fragments, with 83.91% average polymorphism and with an average of 14.10 bands per primer. The size of amplicons varied from 200 to 2500 bp. While, ISSR primers produced 203 fragments of which 161 were polymorphic with an average of 11.28 bands per primer with 73.25% average polymorphism. The size of amplicons ranges from 200 to 2500 bp. RAPD and ISSR markers were also assessed by calculating polymorphic information content (PIC) to discriminate the genotypes; the average PIC value for RAPD, ISSR, and combined RAPD + ISSR markers obtained was more than 0.50 suggesting the informativeness of markers. UPGMA analysis based on Jaccard’s similarity coefficient for RAPD, ISSR, and RAPD + ISSR data reveals that 40 accessions of E. ribes were depicted in four clusters. The clustering pattern of all individuals in PCoA analysis agreed with the UPGMA dendrograms, which further confirms the genetic relationships explained by cluster analysis. AMOVA analysis of RAPD, ISSR, and combined marker system revealed variation within the population, ranging from 41 to 44%, and among the population, it ranged from 56 to 59%.
Conclusion
The present study provides an optimized method for evaluating the genetic diversity of Embelia ribes using RAPD and ISSR markers which are useful for further sustainable utilization and conservation of natural populations in the Western Ghats of India.



Similar content being viewed by others
References
Angiosperm Phylogeny Group IV (APG IV) (2016) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181:1–20. https://doi.org/10.1111/boj.12385
Kamble V, Attar U, Umdale S, Nimbalkar M, Ghane S, Gaikwad N (2020) Phytochemical analysis, antioxidant activities and optimized extraction of embelin from different genotypes of Embelia ribes Burm f.: a woody medicinal climber from Western Ghats of India. Physiol Mol Biol Plants 26(9):1855–1865
Chitra M, Sukumar E, Suja V, Devi S (1994) Antitumor, anti-inflammatory and analgesic property of embelin, a plant product. Chemotherapy 40:109–113
Khan S, Balick MJ (2001) Therapeutic plants of Ayurveda: a review of selected clinical and other studies for 166 species. J Altern Complement Med 7:405–515
Bhandari U, Kanojia R, Pillai KK (2002) Effect of ethanolic extract of Embelia ribes on dyslipidemia in diabetic rats. Int J Exp Diabetes Res 3:159–162
Xu M, Cui J, Fu H et al (2005) Embelin derivatives and their anticancer activity through microtubule disassembly. Planta Med 71:944–948
Ansari MN, Bhandari U (2008) Antihyperhomocysteinemic activity of an Ethanol Extract from Embelia ribes. albino Rats Pharm Biol 46:283–287
Ansari MN, Bhandari U (2008) Effect of an ethanol extract of Embelia ribes fruits on isoproterenol-induced myocardial infarction in albino rats. Pharm Biol 46:928–932
Singh D, Singh R, Singh P, Gupta RS (2009) Effects of embelin on lipid peroxidation and free radical scavenging activity against liver damage in rats. Basic Clin Pharmacol Toxicol 105:243–248
Chaudhari HS, Bhandari U, Khanna G (2012) Preventive effect of embelin from embelia ribes on lipid metabolism and oxidative stress in high-fat diet-induced obesity in rats. Planta Med 78:651–657
Durg S, Veerapur VP, Neelima S, Dhadde SB (2017) Antidiabetic activity of Embelia ribes, embelin and its derivatives: A systematic review and meta-analysis. Biomed Pharmacother 86:195–204
Ravikumar K, Ved DK, Vijaya Sankar R, Udayan PS (2000) 100 Red listed medicinal plants of conservation concern in Southern India
Rajashekaran PE (2001) Biodiversity of threatened species of medicinal plants in India. Trends Wildl Biodivers Conserv Manag 2:104–125
Myers N (1988) Threatened biotas:“ hot spots” in tropical forests. Environmentalist 8:187–208
Grover A, Sharma PC (2016) Development and use of molecular markers: past and present. Crit Rev Biotechnol 362:290–302
Petit RJ, Hampe A (2006) Some evolutionary consequences of being a tree. Annu Rev Ecol Evol Syst 37:187–214. https://doi.org/10.1146/annurev.ecolsys.37.091305.110215
Devaiah KM, Venkatasubramanian P (2008) Genetic characterization and authentication of Embelia ribes using RAPD-PCR and SCAR marker. Planta Med 74:194–196
Nagamani V, Rani AS Development of RAPD Markers for Identification and Authentification of Embelia ribes ARed Listed Indian Medicinal Plant
Gowda B, Chandrika K, Prasanna K, Kirana V (2010) AFLP authentication of Embelia ribes Burm. f. and Embelia tsjeriam-cottam A. DC. Int J Sci Nat 1(1):58–60
Chrungoo NK, Rout GR, Balasubramani SP et al (2018) Establishing taxonomic identity and selecting genetically diverse populations for conservation of threatened plants using molecular markers.Curr Sci539–553
Prajapat P, Sasidharan N, Ballani A (2015) Assessment of genetic diversity in four brassica species using randomly amplifed polymorphic DNA markers. Int J Agric Environ Biotechnol 84:831–836
Bhattacharyya P, Kumaria S (2015) Molecular characterization of Dendrobium nobile Lindl., an endangered medicinal orchid, based on randomly amplifed polymorphic DNA. Plant Syst Evol 3011:201–210
Salazar-Laureles ME, Pérez López DDJ, González- Huerta A, Vázquez-García LM, Valadez-Moctezuma E (2015) Genetic variability analysis of faba bean accessions using Inter-simple sequence repeat (ISSR) markers. Chil J Agric Res https://doi.org/10.4067/S0718-58392015000100017
Asfaw BM, Dagne K, Keneni G, Kemal S, Kassahu T (2018) Genetic diversity study of Ethiopian Faba bean (Vicia faba L.) varieties based on phenotypic traits and inter simple sequence repeat (ISSR) markers. Afr J Biotechnol. https://doi.org/10.5897/AJB2017.16331
Kapteyn J, Goldsbrough P, Simon J (2002) Genetic relationships and diversity of commercially relevant Echinacea species. Theor Appl Genet 105:369–376
Moghaieb REA, Abdelhadi AA, El-Sadawy HA et al (2017) Molecular identification and genetic diversity among Photorhabdus and Xenorhabdus isolates. 3 Biotech 7:1–9
Verma KS, Kachhwaha S, Kothari SL (2013) In vitro plant regeneration of Citrullus colocynthis (L.) Schard. and assessment of genetic fidelity using ISSR and RAPD markers. Indian J Biotech 12:409–414
Khurana-Kaul V, Kachhwaha S, Kothari SL (2012) Characterization of genetic diversity in Jatropha curcas L. germplasm using RAPD and ISSR markers. Indian J Biotech 11:54–61
Velasco-Ramírez AP, Torres-Morán MI, Molina-Moret S et al (2014) Efficiency of RAPD, ISSR, AFLP and ISTR markers for the detection of polymorphisms and genetic relationships in camote de cerro (Dioscorea spp.). Electron J Biotechnol 17:65–71
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bull 19:11–15
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA (1990) Nucleic Acids, DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nuc Res 18:6531–6535
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Rohlf FJ (1998) NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.1. Exeter Software, Setauket, New York
Sneath PHA, Sokal RR (1973) Numerical taxonomy: the principles and practice of numerical classification. San Francisco: Freeman, 1973. 573 p
Hammer O, Harper D, Ryan PD (2001) Past: paleontological statistics software package for education and data analysis. palaeontol electron 4:9
Liu K, Muse SV (2005) Power marker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129. doi:https://doi.org/10.1093/bioinformatics/bti282
Smith JSC, Chin ECL, Shu H et al (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theor Appl Genet 95:163–173
Prevost A, Wilkinson M (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Varshney RK, Chabane K, Hendre PS, Aggarwal RK, Graner A (2007) Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci 173:638–649
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel, Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Tamboli AS, Yadav PB, Gothe AA, Yadav SR, Govindwar SP (2018) Molecular phylogeny and genetic diversity of genus Capparis (Capparaceae) based on plastid DNA sequences and ISSR markers. Plant Syst Evol 304(2):205–217. https://doi.org/10.1007/s00606-017-1466-z
Vinceti B, Loo J, Gaisberger H, Van Zonneveld MJ, Schueler S, Konrad H (2013) Conservation priorities for Prunus africana defned with the aid of spatial analysis of genetic data and climatic variables. PLoS ONE 8(3):e59987
Stevens L, Salomon B, Sun G (2007) Microsatellite variability and heterozygote excess in Elymus trachycaulus populations from British Columbia in Canada. Biochem Syst Ecol 35:725–736. doi:https://doi.org/10.1016/j.bse.2007.05.017
Shrisha NB, Kigga KSK, Ramachandra KK (2018) Genetic diversity of Embelia species, Maesa indica and Ardisia solanacea sampled from Western Ghats of Karnataka using DNA markers Res J Life Sci Bioinform. Pharma Chem Sci 4(6):457–467
Jayaram K, Prasad MNV (2008) Genetic diversity in Oroxylum indicum L. Vent. Bignoniaceae, a vulnerable medicinal plant by random amplifed polymorphic DNA marker. Afr J Biotech 7:254–26265
Pei YL, Zou YP, Yin Z, Wang XQ, Zhang ZX, Hong DY (1995) Preliminary report of RAPD analysis in Paeonia sufruticosa subsp. spontanea and P. rockii. Acta Phys Sin 33:350–35666
Brauner S, Crawford DJ, Stuessy TF (1992) Ribosomal DNA and RAPD variation in the rare plant family Lactoridaceae. Am J Bot 79:1436–143967
Abedian M, Talebi M, Golmohammdi HR, Sayed-Tabatabaei BE (2012) Genetic diversity and population structure of mahaleb cherry (Prunus mahaleb L.) and sweet cherry (Prunus avium L.) using SRAP markers. Biochem Syst Ecol 40:112–117. doi:https://doi.org/10.1016/j.bse.2011.10.005
TilwariA, Sharma R (2021) Random amplifed polymorphic DNA and inter simple sequence repeat markers reveals genetic diversity between micro propagated, wild and feld cultivated genotypes of Gloriosa superba: an endangered medicinal plant. Mol Biology Rep 48:2437–2452
Farajpour M, Ebrahimi M, Amiri R et al (2011) Study of genetic variation in yarrow using inter-simple sequence repeat (ISSR) and random amplifed polymorphic DNA (RAPD) markers. Afr J Biotechnol 10(54):11137–1114174
Patel DM, Fougat RS, Sakure AA, Kumar S, Kumar M, Mistry JG (2016) Detection of genetic variation in sandalwood using various DNA markers. 3 Biotech 61:55
Velasco-Ramirez AP, Torres-Moran MI, Molina-Moret S et al (2014) Efciency of RAPD, ISSR, AFLP and ISTR markers for the detection of polymorphisms and genetic relationships in camote de cerro Dioscorea spp. Electron J Biotechnol 172:65–71. https://doi.org/10.1016/j.ejbt.2014.01.00
Debajit S, Sukriti D, Sneha G, Mohan L et al (2015) RAPD and ISSR based Intra-specifc molecular genetic diversity analysis of Cymbopogon fexuosus L. Stapf with a distinct correlation of morpho-chemical observations. Res J Biotechnol 107:105–113
Acknowledgements
The authors thank the Head, Department of Botany, Shivaji University, Kolhapur, for providing the necessary facilities. We also thank the Maharashtra and Karnataka State Biodiversity Board for providing the necessary permission. VVK is thankful to Mr. V. Y. Kamble, Ganga, Vinayak, and Sushant Sawant for the collections. VVK is thankful to UGC, New Delhi, for financial support under UGC-BSR Fellowship in Sciences for Students (No.F.25 − 1/2013-14 (BSR)/7-163/2007(BSR). Thanks are due to the Natural National Science Foundation of China (NSFC Grant Number 32250410305) awarded to SAR.
Author information
Authors and Affiliations
Contributions
VVK and NBG designed the study. NBG supervised the study. VVK, NBG, and SDU collected the samples. VVK conducted laboratory experiments. SAR, VVK, AST, and SDU conducted bioinformatics and statistical analyses. SAR, VVK, SHW, HL, and AST drafted and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors Vidya V. Kamble and Shabir A. Rather have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.










Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kamble, V.V., Tamboli, A.S., Umdale, S.D. et al. Evaluating genetic diversity of geographically diverse populations of Embelia ribes Burm f., a highly medicinal woody liana from the Western Ghats of India, using random amplified polymorphic DNA (RAPD) and intersimple sequence repeats (ISSR) markers. Mol Biol Rep 50, 1603–1615 (2023). https://doi.org/10.1007/s11033-022-08099-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08099-1