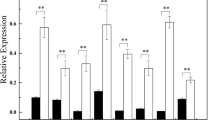
Abstract
Background
Sugarcane is an important industrial plant cultivated mostlyin the arid and semiarid regions. Due to climate change and anthropogenic activities, the sugarcane fieldsare prone to be damagedas a result of salt deposition. The consequence of such phenomena is turning to become a major thread in sugarcane cultivation. To address this issue, the identification of salinity tolerant cultivars would be a suitable strategy to minimize yield loss in the area. It is well known thatthe expression of abiotic stress-responsive genes including noncoding microRNAs (miRNAs) and their codingtargetscould lead to enhancement of stress tolerance in crops. Therefore, the expression study of those noncoding and coding genes under stress conditions is an appropriate approach to screen the tolerant cultivars. In addition, the examination of the expression of miRNA’s target genes could provide deeper insight into the molecular stress mechanism and facilitate the identification of tolerant cultivars.
Methods and results
We aimedto assess the expression of nine candidate miRNAsand their corresponding targeted genes among the studied sugarcane cultivars under high salinity conditions, leading to the identification of the salt-tolerant cultivar. To achieve our goal, a two-factorial experiment with three sugarcane cultivars (CP-48, CP-57, CP-69) and two salinity levels (0 and 8 ds/m) was conducted. The result indicated significant differences in expression with in miRNAs and also their target genes. The highest reduction of miRNAs expression occurred in miR160 while the lowest oneappeared in miR1432. The data also indicated that the higher and the lowest expression of targeted genes occurred in miR160 and miR393 respectively. Among studied cultivars, the CP-57 showed poor performance while CP-69 expresses a superior tolerance to salt stress.
Conclusions
Taken together, these results suggested that the monitoring of microRNA expressioncould provide a new approach forthe screening of well-adapted cultivars under salt conditions. Such an approach would be the appropriate solutionto combat plant stress inhigh salinity region/soil. Our result indicated that the miR160 generates sugarcane tolerant to salt stress, can be potentially be used as a biomarker to salt stress.




Similar content being viewed by others
References
Zhang M, Govindaraju M (2018) Sugarcane production in China. In de Oliveira, A. B. (ed) Sugarcane-technology and research, 1stedn. IntechOpen, London, pp 49-68. https://doi:10.5772/intechopen.73113
Hosseini-Zare N, Gholami A, Panahpour E, Jafarnejadi A (2014) Pollution load assessment in the soil and water resources: a case study in Karun river drainage basin, southwest of Iran. Online J Nat 3(3):427.
Ahmadi E, McLellan B, Mohammadi-Ivatloo B, Tezuka T (2020) The role of renewable energy resources in sustainability of water desalination as a potential fresh-water source: An updated review.Sustainability. 12(13):5233. https://doi.org/10.3390/su12135233
Hussain N, Ghaffar A, Zafar ZU, Javed M, Shah KH, Noreen S, Manzoor H, Iqbal M, Hassan IF, Bano H, Gul HS (2021) Identification of novel source of salt tolerance in local bread wheat germplasm using morpho-physiological and biochemical attributes. Sci Rep 11(1):1–2. https://doi: 10.1038/s41598-021-90280-w.
Zhao C, Zhang H, Song C, Zhu JK, Shabala S (2020) Mechanisms of plant responses and adaptation to soil salinity. Innovation (Camb) 1(1):100017. https://doi:10.1016/j.xinn.2020.100017
Zhang B, Wang Q (2015) MicroRNA-based biotechnology for plant improvement. J Cell Physiol 230(1):1–15. https://doi:10.1002/jcp.24685
Ku YS, Wong JW, Mui Z, Liu X, Hui JH, Chan TF, Lam HM (2015) Small RNAs in plant responses to abiotic stresses: regulatory roles and study methods. Int J Mol Sci 16(10):24532–24554. https://doi:10.3390/ijms161024532
Zhang B (2015) MicroRNA: a new target for improving plant tolerance to abiotic stress. J Exp Bot. 66(7):1749-61. https://doi:10.1093/jxb/erv013
Bhatnagar-Mathur P, Vadez V, Sharma Kiran K (2008) Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep 27(3):411-24. https://doi:10.1007/s00299-007-0474-9
Bottino MC, Rosario S, Grativol C, Thiebaut F, Rojas CA, Farrineli L, Hemerly AS, Ferreira PC (2013) High-throughput sequencing of small RNA transcriptome reveals salt stress regulated microRNAs in sugarcane. PLoS ONE 8(3): e59423. https://doi.org/10.1371/journal.pone.0059423
Sunkar R, Li Y-F, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17(4):196–203. https://doi:10.1016/j.tplants.2012.01.010.
Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD, Morris Q (2013) GeneMANIA prediction server 2013 update. Nucleic Acids Res 41(W1):W115–W122. https://doi:10.1093/nar/gkt533
Deng W, Wang Y, Liu Z, Cheng H, Xue Y (2014) HemI: a toolkit for illustrating heatmaps. PLoS One 9(11):e111988. https://doi.org/10.1371/journal.pone.0111988
Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant methods 3(1):1–2. https://doi.org/10.1186/1746-4811-3-12
Lin JS, Kuo CC, Yang IC, Tsai WA, Shen YH, Lin CC, Liang YC, Li YC, Kuo YW, King YC, Lai HM (2018) MicroRNA160 modulates plant development and heat shock protein gene expression to mediate heat tolerance in Arabidopsis. Front Plant Sci 19:68. https//doi:10.3389/fpls.2018.00068
Xu J, Li J, Cui L, Zhang T, Wu Z, Zhu PY, Meng YJ, Zhang KJ, Yu XQ, Lou QF, Chen JF (2017) New insights into the roles of cucumber TIR1 homologs and miR393 in regulating fruit/seed set development and leaf morphogenesis. BMC Plant Biol 17(1):1–4. https://doi.org/10.1186/s12870-017-1075-6
Djami-Tchatchou AT, Sanan-Mishra N, Ntushelo K, Dubery IA (2017) Functional roles of microRNAs in agronomically important plants potential as targets for crop improvement and protection. Front. Plant Sci 8:378. https://doi.org/10.3389/fpls.2017.00378
Lee SH, Li CW, Koh KW, Chuang HY, Chen YR, Lin CS, Chan MT (2014) MSRB7 reverses oxidation of GSTF2/3 to confer tolerance of Arabidopsis thaliana to oxidative stress. J Exp Bot 65(17):5049–5062. https://doi:10.1093/jxb/eru270
Dastidar MG, Scarpa A, Mägele I, Ruiz-Duarte P, von Born P, Bald L, Jouannet V, Maizel A (2019) ARF5/MONOPTEROS directly regulates miR390 expression in the Arabidopsis thaliana primary root meristem. Plant direct 3(2):e00116. https://doi: 10.1002/pld3.116
Bouzroud S, Gouiaa S, Hu N, Bernadac A, Mila I, Bendaou N, Smouni A, Bouzayen M, Zouine M (2018) Auxin response factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PloS one13(2):e0193517. https://doi:10.1371/journal.pone.0193517
Xu X, Yin L, Ying Q, Song H, Xue D, Lai T, Xu M, Shen B, Wang H, Shi X (2013) High-throughput sequencing and degradome analysis identify miRNAs and their targets involved in fruit senescence of Fragaria ananassa. PLoS ONE 8(8):e70959. https://doi.org/10.1371/journal.pone.0070959
Dudhate A, Shinde H, Yu P, Tsugama D, Gupta SK, Liu S, Takano T (2021) Comprehensive analysis of NAC transcription factor family uncovers drought and salinity stress response in pearl millet (Pennisetum glaucum). BMC Genom 22(1):1–5. https://doi.org/10.1186/s12864-021-07382-y
Zhai L, Liu Z, Zou X, Jiang Y, Qiu F, Zheng Y, Zhang Z (2013) Genome-wide identification and analysis of microRNA responding to long‐term waterlogging in crown roots of maize seedlings. Physiol Plant 147(2):181–193. https://doi:10.1111/j.1399-3054.2012.01653.x.
Sun G (2012) MicroRNAs and their diverse functions in plants. Plant Mol Biol 80(1):17–36. https://doi:10.1007/s11103-011-9817-6
Ó’Maoiléidigh DS, van Driel AD, Singh A, Sang Q, Le Bec N, Vincent C, de Olalla EB, Vayssières A, Romera Branchat M, Severing E, Martinez Gallegos R (2021) Systematic analyses of the MIR172 family members of Arabidopsis define their distinct roles in regulation of APETALA2 during floral transition. PLoS Biol 19(2):e3001043. https://doi:10.1371/journal.pbio.3001043
Gazon H, Barbeau B, Mesnard JM, Peloponese JM Jr (2018) Hijacking of the AP-1 Signaling Pathway during Development of ATL. Front microbiol 15:8:2686. https://doi.org/10.3389/fmicb.2017.02686
Wen FL, Yue Y, He TF, Gao XM, Zhou ZS, Long XH (2020) Identification of miR390-TAS3-ARF pathway in response to salt stress in Helianthus tuberosus L. Gene 15:738:144460. https://doi: 10.1016/j.gene.2020.144460
Chen ZH, Bao ML, Sun YZ, Yang YJ, Xu XH, Wang JH, Han N, Bian HW, Zhu MY (2011) Regulation of auxin response by miR393-targeted transport inhibitor response protein1 is involved in normal development in Arabidopsis. Plant Mol Biol 77(6):619–629. https://doi: 10.1007/s11103-011-9838-1
Ling LZ, Zhang SD (2012) Exploring the evolutionary differences of SBP-box genes targeted by miR156 and miR529 in plants. Genetica 140(7–9):317–324. https://doi: 10.1007/s10709-012-9684-3
Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J (2012) The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193(4):842–851. https://doi: 10.1111/j.1469-8137.2011.04002.x
Mangrauthia SK, Agarwal S, Sailaja B, Madhav MS, Voleti SR (2013) MicroRNAs and their role in salt stress response in plants. In: Ahmad P, Azooz M M, Prasad M N V (ed) Salt stress in plants, 1st edn. Springer, New York, pp 15–46. https://doi:10.1007/978-1-4614-6108-1_2
Acknowledgements
We thank Dr. H. Hamdi the research center chief of sugarcane development, Ahvaz, Iran, for the donation of sugarcanes seeds.
Author information
Authors and Affiliations
Contributions
Writing–original draft and investigation:Tofigh Mazalmazraei; Supervision andproject administration: Leila Nejhad sadeghi; Data curation and formal analysis: Khosro Mehdi Khanlou; Validation: Daryoosh Nabati Ahmadi.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflicts of interes.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.

Rights and permissions
About this article
Cite this article
Mazalmazraei, T., Nejadsadeghi, L., Mehdi Khanlou, K. et al. Comparative analysis of differentially expressed miRNAs in leaves of three sugarcanes (Saacharum officinarum L.) cultivars during salinity stress. Mol Biol Rep 50, 485–492 (2023). https://doi.org/10.1007/s11033-022-07349-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07349-6