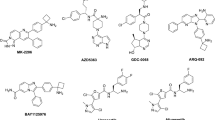
Abstract
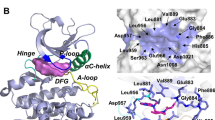
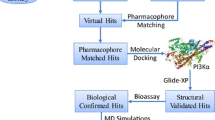
Akt1, as an important member of the Akt family, plays a controlled role in cancer cell growth and survival. Inhibition of Akt1 activity can promote cancer cell apoptosis and inhibit tumor growth. Therefore, in this investigation, a multilayer virtual screening approach, including receptor–ligand interaction-based pharmacophore, 3D-QSAR, molecular docking, and deep learning methods, was utilized to construct a virtual screening platform for Akt1 inhibitors. 17 representative compounds with different scaffolds were identified as potential Akt1 inhibitors from three databases. Among these 17 compounds, the Hit9 exhibited the best inhibitory activity against Akt1 with inhibition rate of 33.08% at concentration of 1 μM. The molecular dynamics simulations revealed that Hit9 and Akt1 could form a compact and stable complex. Moreover, Hit9 interacted with some key residues by hydrophobic, electrostatic, and hydrogen bonding interactions and induced substantial conformation changes in the hinge region of the Akt1 active site. The average binding free energies for the Akt1-CQU, Akt1-Ipatasertib, and Akt1-Hit9 systems were − 34.44, − 63.37, and − 39.14 kJ mol−1, respectively. In summary, the results obtained in this investigation suggested that Hit9 with novel scaffold may be a promising lead compound for develo** new Akt1 inhibitor for treatment of various cancers with Akt1 overexpressed.
Graphical abstract











Similar content being viewed by others
Data availability
The data presented in this study are available on request from the corresponding author.
References
Shariati M, Meric-Bernstam F (2019) Targeting AKT for cancer therapy. Expert Opin Inv Drug 28(11):977–988. https://doi.org/10.1080/13543784
Franke T (2008) PI3K/Akt: getting it right matters. Oncogene 27(50):6473–6488. https://doi.org/10.1038/onc.2008.313
**ng Y, Lin NU, Maurer M, Huiqin C, Armeen M (2019) Phase II trial of AKT inhibitor MK-2206 in patients with advanced breast cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN loss/PTEN mutation. Breast Cancer Res 21:1–12. https://doi.org/10.1186/s13058-019-1154-8
Yang ZZ, Tschopp O, Hemmings-Mieszczak M (2003) Protein kinase Bα/Akt1 regulates placental development and fetal growth. J Biol Chem 278(34):32124–32131
Rao G, Pierobon M, Kim IK (2017) Inhibition of AKT1 signaling promotes invasion and metastasis of non-small cell lung cancer cells with K-RAS or EGFR mutations. Sci Rep 7(1):1–12. https://doi.org/10.1038/s41598-017-06128-9
Song M, Bode AM, Dong Z (2019) AKT as a therapeutic target for cancer challenging cancer therapy by targeting AKT. Cancer Re 79(6):1019–1031
Manning BD, Cantley LC (2007) AKT/PKB signaling: navigating downstream. Cell 129(7):1261–1274. https://doi.org/10.1016/j.cell.2007.06.009
Manning BD, Toker A (2017) AKT/PKB signaling: navigating the network. Cell 169(3):381–405. https://doi.org/10.1016/j.cell.2017.04.001
Cheng JQ, Lindsley CW, Cheng G (2005) The Akt/PKB pathway: molecular target for cancer drug discovery. Oncogene 24(50):7482–7492. https://doi.org/10.1038/sj.onc.1209088
Bono JS, Giorgi UD, Rodrigues DN (2019) Randomized phase II study evaluating Akt blockade with -ipatasertib, in combination with abiraterone, in patients with metastatic prostate cancer with and without PTEN loss-ipatasertib in prostate cancer with and without PTEN loss. Clin Cancer Res 25(3):928–936. https://doi.org/10.1158/1078-0432
Hirai H, Sootome H, Nakatsuru Y (2010) MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivoMK-2206 sensitizes tumors to chemotherapy. Mol Cancer Ther 9(7):1956–1967
Yu Y, Savage RE, Eathiraj S (2015) Targeting AKT1-E17K and the PI3K/AKT pathway with an allosteric AKT inhibitor, ARQ 092. PLoS ONE 10(10):e0140479. https://doi.org/10.1371/journal.pone.0140479
Politz O, Siegel F, Bärfacker L (2017) BAY 1125976, a selective allosteric AKT1/2 inhibitor, exhibits high efficacy on AKT signaling-dependent tumor growth in mouse models. Int J Cancer 140(2):449–459. https://doi.org/10.1002/ijc.30457
Weisner J, Landel I, Reintjes C (2019) Preclinical efficacy of covalent-allosteric AKT inhibitor borussertib in combination with trametinib in KRAS-mutant pancreatic and colorectal cancer preclinical efficacy of AKT inhibitor borussertib. Cancer Res 79(9):2367–2378. https://doi.org/10.1158/0008-5472
Baron R, Baron R (2012) Computational drug discovery and design. Springer, New York. https://doi.org/10.3390/molecules25061375
Anderson AC (2003) The process of structure-based drug design. Int J Mol Sci 10(9):787–797. https://doi.org/10.1016/j.chembiol.2003.09.002
Singh N, Chaput L, Villoutreix BO (2021) Virtual screening web servers: designing chemical probes and drug candidates in the cyberspace. Brief Bioinform 22(2):1790–1818. https://doi.org/10.1093/bib/bbaa034
Lin X (2020) A reviews on applications of computational methods in drug screening and design. Molecules 25(6):1375. https://doi.org/10.3390/molecules25061375
Dsouza S, Prema K, Balaji S (2020) Machine learning models for drug–target interactions: current knowledge and future directions. Drug Discov Today 25(4):748–756. https://doi.org/10.1016/j.drudis.2020.03.003
Mei D, Yin Y, Wu F (2015) Discovery of potent and selective urea-based ROCK inhibitors: exploring the inhibitor’s potency and ROCK2/PKA selectivity by 3D-QSAR, molecular docking and molecular dynamics simulations. Biomedicines 23(10):2505–2517. https://doi.org/10.1016/j.bmc.2015.03.047
Shaik NA, Hakeem KR, Banaganapalli B (2019) Essentials of bioinformatics, vol II. Springer, New York
Alder BJ, Wainwright TE (1957) Phase transition for a hard sphere system. J Chem Phys 27(5):1208–1209
Morris GM, Goodsell DS, Halliday RS (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Ewing TJ, Makino S, Skillman AG (2001) DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des 15:411–428. https://doi.org/10.1023/a:1011115820450
Inc A (2010) Discovery studio 3.5 help. Accelrys Software Inc., San Diego
Trott O, Olson A (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Gaillard T (2018) Evaluation of AutoDock and AutoDock Vina on the CASF-2013 benchmark. J Chem Inf Model 58(8):1697–1706. https://doi.org/10.1021/acs.jcim.8b00312
Rao SN, Head MS, Kulkarni A (2007) Validation studies of the site-directed docking program LibDock. J Chem Inf Model 47(6):2159–2171. https://doi.org/10.1021/ci6004299
Wu G, Robertson DH, Brooks CL (2003) Detailed analysis of grid-based molecular docking: a case study of CDOCKER—a CHARMm-based MD docking algorithm. J Comput Chem 24(13):1549–1562. https://doi.org/10.1002/jcc.10306
Lippa B, Pan G, Corbett M (2008) Synthesis and structure based optimization of novel Akt inhibitors. Bioorg Med Chem Lett 18(11):3359–3363. https://doi.org/10.1016/j.bmcl.2008.04.034
Babu S, Nagarajan S, Sathish S, Negi V, Sohn H, Madhavan T (2022) Identification of potent and selective JAK1 lead compounds through ligand-based drug design approaches. Front Pharmacol. https://doi.org/10.3389/fphar.2022.837369
Cramer RD, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967. https://doi.org/10.1002/chin.198851058
Liu Z, Du J, Fang J (2019) DeepScreening: a deep learning-based screening web server for accelerating drug discovery. Database. https://doi.org/10.1093/database/baz104
**ong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A, Chen X, Hou T, Cao D (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res 49(W1):W5–W14. https://doi.org/10.1093/nar/gkab255
Bickerton GR, Paolini GV, Besnard J (2012) Quantifying the chemical beauty of drugs. Nat Chem 4(2):90–98. https://doi.org/10.1038/nchem.1243
Berendsen H, Hess B, Lindahl E (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718. https://doi.org/10.1002/jcc.20291
Tian C, Kasavajhala K, Belfon K (2019) Ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J Chem Theory Comput 16(1):528–552. https://doi.org/10.1021/acs.jctc.9b00591
Mcgibbon R, Beauchamp K, Harrigan M (2015) MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys J 109(8):1528–1532. https://doi.org/10.1016/j.bpj.2015.08.015
Grant B, Skjærven L, Yao X (2021) The Bio3D packages for structural bioinformatics. Protein Sci 30(1):20–30. https://doi.org/10.1002/pro.3923
Kumari R, Kumar R, Consortium O (2014) A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962. https://doi.org/10.1021/ci500020m
**e J, Meng D, Li Y (2022) Virtual screening for potential discoidin domain receptor 1 (DDR1) inhibitors based on structural assessment. Mol Divers. https://doi.org/10.1007/s11030-022-10557-8
Fratev F, Gutierrez DA, Aguilera RA, Sirimulla S (2021) Discovery of new AKT1 inhibitors by combination of in silico structure based virtual screening approaches and biological evaluations. J Biomol Struct Dyn 39(1):368–377. https://doi.org/10.26434/chemrxiv.7591202.v1
Liu Y, Yin Y, Zhang Z (2017) Structural optimization elaborates novel potent Akt inhibitors with promising anticancer activity. Eur J Med Chem 29(138):543–551. https://doi.org/10.1016/j.ejmech.2017.06.067
Dong X, Zhan W, Zhao M (2019) Discovery of 3,4,6-trisubstituted piperidine derivatives as orally active, low hERG blocking Akt inhibitors via conformational restriction and structure-based design. J Med Chem 62(15):7264–7288. https://doi.org/10.1021/acs.jmedchem.9b00891
Sadeghi F, Afkhami A, Madrakian T, Ghavami R (2021) Computational study on subfamilies of piperidine derivatives: QSAR modelling, model external verification, the inter-subset similarity determination, and structure-based drug designing. SAR QSAR Environ Res 32(6):433–462. https://doi.org/10.1080/1062936X.2021.1891568
Zeng Q, Bourbeau MP, Wohlhieter GE, Yao G (2010) 2-Aminothiadiazole inhibitors of AKT1 as potential cancer therapeutics. Bioorg Med Chem Lett 20(5):1652–1656. https://doi.org/10.1016/j.bmcl.2010.01.046
Rice KD, Kim MH, Bussenius J (2012) Pyrazolopyrimidines as dual Akt/p70S6K inhibitors. Bioorg Med Chem Lett 22(8):2693–2697. https://doi.org/10.1016/j.bmcl.2012.03.011
**ao Y, Huck BR, Lan R (2021) Discovery of 4-aminopyrimidine analogs as highly potent dual P70S6K/Akt inhibitors. Bioorg Med Chem Lett 15(50):128352. https://doi.org/10.1016/j.bmcl.2021.128352
Zhan W, Che J, Xu L (2019) Discovery of pyrazole-thiophene derivatives as highly potent, orally active Akt inhibitors. Eur J Med Chem 15(180):72–85. https://doi.org/10.1016/j.ejmech.2019.07.017
Zhan W, Xu L, Dong X (2016) Design, synthesis and biological evaluation of pyrazol-furan carboxamide analogues as novel Akt kinase inhibitors. Eur J Med Chem 19(117):47–58. https://doi.org/10.1016/j.ejmech.2016.03.074
Funding
Open access funding provided by the National Natural Science Foundation of China (Grant No. 82260693) and Science and Technology Program Project of Gansu Province (20JR5RA538).
Author information
Authors and Affiliations
Contributions
WZ Investigation, Methodology, Writing-original draft; M-LH Investigation, Validation; X-YS Validation, Formal analysis; X-LC Validation, Data curation; XS and H-ZQ Methodology, Validation; YL Funding acquisition, Project administration; HZ Conceptualization, Writing – review & editing, Project administration, Investigation, Supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent to participate
Consent to participate was obtained from all the authors.
Consent for publication
Consent for publication was obtained from all the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, W., Hu, ML., Shi, XY. et al. Discovery of novel Akt1 inhibitors by an ensemble-based virtual screening method, molecular dynamics simulation, and in vitro biological activity testing. Mol Divers (2024). https://doi.org/10.1007/s11030-023-10788-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10788-3