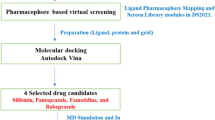
Abstract
Drug repurposing opens new avenues in cancer therapy. Drug repurposing, or finding new uses for existing drugs, can substantially reduce drug discovery time and costs. Cheminformatics, genetics, and systems biology advances enable repositioning drugs. Clinical usage of PD-1/PD-L1 blocking has been approved because of its efficacy in improving prognosis in select groups. The PD-1/PD-L1 axis was considered to represent a mechanism for tumour evasion of host tumour antigen-specific T-cell immunity in early preclinical research. The expression of PD-L1 in cancer cells causes T lymphocytes to become exhausted by transmitting a co-inhibitory signal. A better understanding of how PD-L1 is regulated in cancer cells could lead to new therapeutic options. In this view, the study was aimed to repurpose the existing FDA-approved drugs as a potential PD-L1 inhibitor through e-Pharmacophore modelling, molecular docking and dynamic simulation. e-Pharmacophore screening retrieved 324 FDA-approved medications with the fitness score ≥ 1. The top 10-docked FDA candidates were compared with IN-35 (Clinical trial candidate) for its interaction pattern with critical amino acid residues. Mirabegron and Indacaterol exhibited a greater affinity for PD-L1 with docking scores of − 9.213 kcal mol−1 and − 8.023 kcal mol−1, respectively. Mirabegron retain interactions at all three major hotspots in the PD-L1 dimer interface similar to IN-35. MM-GBSA analyses indicated that Mirabegron uses less energy to create a more stable complex and retains all of the inhibitor’s positive interactions found in clinical trial ligand IN-35. Molecular dynamics simulation analysis of the Mirabegron complex showed a similar pattern of deviation in correlation with IN-35, and it retains the interaction with the active key amino acids throughout the simulation time. Our present study has shown Mirabegron as a powerful inhibitor of PD-L1 expression in cancer cells using a drug-repurposing screen.
Graphical abstract









Similar content being viewed by others
References
Cha JH, Chan LC, Li CW, Hsu JL, Hung MC (2019) Mechanisms controlling PD-L1 expression in cancer. Mol Cell 76(3):359–370. https://doi.org/10.1016/j.molcel.2019.09.030
Akinleye A, Rasool Z (2019) Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J Hematol Oncol 12(1):92. https://doi.org/10.1186/s13045-019-0779-5
Sun C, Mezzadra R, Schumacher TN (2018) Regulation and function of the PD-L1 checkpoint. Immunity 48(3):434–452. https://doi.org/10.1016/j.immuni.2018.03.014
Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, Sica GL (2017) Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355(6332):1423–1427. https://doi.org/10.1126/science.aaf0683
Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, Vale RD (2017) T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 355(6332):1428–1433. https://doi.org/10.1126/science.aaf1292
Ribas A (2012) Tumor immunotherapy directed at PD-1. N Engl J Med 366(26):2517–2519. https://doi.org/10.1056/NEJMe1205943
Francisco LM, Sage PT, Sharpe AH (2010) The PD-1 pathway in tolerance and autoimmunity. Immunol Rev 236:219–242. https://doi.org/10.1111/j.1600-065X.2010.00923.x
Fife BT, Pauken KE, Eagar TN, Obu T, Wu J, Tang Q, Azuma M, Krummel MF, Bluestone JA (2009) Interactions between PD-1 and PD-L1 promote tolerance by blocking the TCR-induced stop signal. Nat Immunol 10(11):1185–1192. https://doi.org/10.1038/ni.1790
Pardoll DM (2012) The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12(4):252–264. https://doi.org/10.1038/nrc3239
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674. https://doi.org/10.1016/j.cell.2011.02.013
Li X, Shao C, Shi Y, Han W (2018) Lessons learned from the blockade of immune checkpoints in cancer immunotherapy. J Hematol Oncol 11(1):31. https://doi.org/10.1186/s13045-018-0578-4
Jiang X, Wang J, Deng X, **ong F, Ge J, **ang B, Wu X, Ma J, Zhou M, Li X, Li Y (2019) Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer 18(1):10. https://doi.org/10.1186/s12943-018-0928-4
Sfanos KS, Bruno TC, Meeker AK, De Marzo AM, Isaacs WB, Drake CG (2009) Human prostate-infiltrating CD8+ T lymphocytes are oligoclonal and PD-1+. Prostate 69(15):1694–1703. https://doi.org/10.1002/pros.21020
Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA (2002) Tumor-associated B7–H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 8(8):793–800. https://doi.org/10.1038/nm730
Ribas A, Wolchok JD (2018) Cancer immunotherapy using checkpoint blockade. Science 359(6382):1350–1355. https://doi.org/10.1126/science.aar4060
Horn L, Spigel DR, Vokes EE, Holgado E, Ready N, Steins M, Poddubskaya E, Borghaei H, Felip E, Paz-Ares L, Pluzanski A (2017) Nivolumab versus docetaxel in previously treated patients with advanced non-small-cell lung cancer: two-year outcomes from two randomized, open-label, phase III trials (CheckMate 017 and CheckMate 057). J Clin Oncol 35(35):3924–3933. https://doi.org/10.1200/JCO.2017.74.3062
Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D (2015) Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 373(2):123–135. https://doi.org/10.1056/NEJMoa1504627
Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, Castellano D (2015) Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med 373(19):1803–1813. https://doi.org/10.1056/NEJMoa1510665
Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F (2015) Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med 373(17):1627–1639. https://doi.org/10.1056/NEJMoa1507643
Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, Worden F (2016) Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 375(19):1856–1867. https://doi.org/10.1056/NEJMoa1602252
Schachter J, Ribas A, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, Larkin J (2017) Pembrolizumab versus ipilimumab for advanced melanoma: final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 390(10105):1853–1862. https://doi.org/10.1016/S0140-6736(17)31601-X
Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK, Necchi A (2017) Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med 376(11):1015–1026. https://doi.org/10.1056/NEJMoa1613683
Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, Von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols MC, Cortinovis DL (2017) Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 389(10066):255–265. https://doi.org/10.1016/S0140-6736(16)32517-X
Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, Park K, Smith D, Artal-Cortes A, Braiteh F, Waterkamp D, He P, Zou W, Yi J, Sandler A, Rittmeyer A (2016) Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387(10030):1837–1846. https://doi.org/10.1016/S0140-6736(16)00587-0
Shen X, Zhao B (2018) Efficacy of PD-1 or PD-L1 inhibitors and PD-L1 expression status in cancer: meta-analysis. BMJ 362:k3529. https://doi.org/10.1136/bmj.k3529
Medina-Franco EL-LJBJL (2021) Informatics for chemistry, biology, and biomedical sciences. J Chem Inform Model 61(1):26–35. https://doi.org/10.1021/acs.jcim.0c01301
Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, Sanseau P, Cavalla D, Pirmohamed M (2019) Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 18(1):41–58. https://doi.org/10.1038/nrd.2018.168
Yella JK, Yaddanapudi S, Wang Y, Jegga AG (2018) Changing trends in computational drug repositioning. Pharmaceuticals (Basel) 11(2):57. https://doi.org/10.3390/ph11020057
Guo X, Ji J, Feng Z, Hou X, Luo Y, Mei Z (2020) A network pharmacology approach to explore the potential targets underlying the effect of sinomenine on rheumatoid arthritis. Int Immunopharmacol 80:106201. https://doi.org/10.1016/j.intimp.2020.106201
Farha MA, Brown ED (2019) Drug repurposing for antimicrobial discovery. Nat Microbiol 4(4):565–577. https://doi.org/10.1038/s41564-019-0357-1
Dotolo S, Marabotti A, Facchiano A, Tagliaferri R (2021) A review on drug repurposing applicable to COVID-19. Brief Bioinform 22(2):726–741. https://doi.org/10.1093/bib/bbaa288
Cheng F, Desai RJ, Handy DE, Wang R, Schneeweiss S, Barabási AL, Loscalzo J (2018) Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat Commun 9(1):2691. https://doi.org/10.1038/s41467-018-05116-5
Verbaanderd C, Meheus L, Huys I, Pantziarka P (2017) Repurposing drugs in oncology: next steps. Trends Cancer 3(8):543–546. https://doi.org/10.1016/j.trecan.2017.06.007
Guo Y, ** Y, Wang B, Liu B (2021) Molecular mechanism of small-molecule inhibitors in blocking the PD-1/PD-L1 pathway through PD-L1 dimerization. Int J Mol Sci 22(9):4766. https://doi.org/10.3390/ijms22094766
Kuang Z, Heng Y, Huang S, Shi T, Chen L, Xu L, Mei H (2020) Partial least-squares discriminant analysis and ensemble-based flexible docking of PD-1/PD-L1 inhibitors: a pilot study. ACS Omega 5:26914–26923. https://doi.org/10.1021/acsomega.0c04149
Acknowledgements
The authors thank the PSG College of Pharmacy, Peelamedu, Coimbatore for providing computational facility.
Author information
Authors and Affiliations
Contributions
JC contributed to the study conception and design. JC, PP and SE performed computational simulation and analysed the data. JC, SE, VM and PP wrote the original manuscript. PT and SK proofread the manuscript. All the authors have read and approved the manuscript for submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
None of the authors conducted any human or animal experiments for this publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chandrasekaran, J., Elumalai, S., Murugesan, V. et al. Computational design of PD-L1 small molecule inhibitors for cancer therapy. Mol Divers 27, 1633–1644 (2023). https://doi.org/10.1007/s11030-022-10516-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10516-3