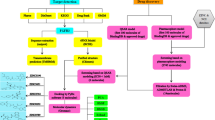
Abstract
Gall bladder cancer (GBC) is an aggressive and most common malignancy of biliary tract lacking effective treatment due to unavailability of suitable biomarkers and therapeutics. SMAD4 is an essential mediator of transforming growth factor-β pathway involved in various cellular processes like growth, differentiation and apoptosis and also recognized as therapeutic target for GBC and other gastrointestinal tract cancers. In the present study, 3D structure of SMAD4 mutants was optimized through molecular dynamics simulation (MDS) along with wildtype. Furthermore, binding site of protein was predicted through hybrid approach and structural based virtual screening against two drug libraries was performed followed by docking. MDS of top docking score protein–ligand complexes were carried, and binding free energy was rescored. Two potential inhibitors, namely ZINC2098840 and ZINC8789167, were screened that displayed higher binding affinity towards mutant proteins compared with wildtype and both hydrophilic as well as hydrophobic interactions play a crucial role during protein–ligand binding. Current study identified novel and potent inhibitors of SMAD4 mutant that could be used as a drug candidate for the development of personalized medicine for gall bladder and other associated cancers.
Graphic abstract









Similar content being viewed by others
Data availability
The datasets supporting the conclusion of this article are included within the article/supplementary material.
Abbreviations
- GBC:
-
Gall bladder cancer
- GROMACS:
-
Groningen machine for chemical simulation
- MDS:
-
Molecular dynamics simulation
- MMD:
-
Million molecular database
- MMPBSA:
-
Molecular mechanics Poisson Boltzmann surface area
- MT:
-
Mutant
- NCD:
-
Natural compound database
- PDB:
-
Protein data bank
- Rg:
-
Radius of gyration
- RMSD:
-
Root-mean-square deviation
- RMSF:
-
Root-mean-square fluctuation
- SAD:
-
SMAD4 activated domain
- SMAD4:
-
Mothers against decapentaplegic homolog 4
- SNP:
-
Single nucleotide polymorphism
- SNV:
-
Single nucleotide variant
- TGF-β:
-
Transforming growth factor-β
- WT:
-
Wildtype
References
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424
MathewGeorgeJKVasudevanJames APSKMDFV (2019) Transition of cancer in populations in India. Cancer Epidemiol 58:111–120
Mehrotra R, Tulsyan S, Hussain S et al (2018) Genetic landscape of gallbladder cancer: global overview. Mutat Res 778:61–71
Kanthan R, Senger JL, Ahmed S, Kanthan SC (2015) Gallbladder cancer in the 21st century. J Oncol 2015:967472
Hundal R, Shaffer EA (2012) Gallbladder cancer: epidemiology and outcome. Clin Epidemiol 6:99–109
Stinton LM, Shaffer EA (2012) Epidemiology of gallbladder disease: cholelithiasis and cancer. Gut Liver 6:172–187
Attisano L, Wrana JL (2002) Signal transduction by the TGF-β superfamily. Science 296:1646–1647
Connolly EC, Freimuth J, Akhurst RJ (2012) Complexities of TGF-β targeted cancer therapy. Int J Biol Sci 8:964
Derynck R, Gelbart WM, Harland RM et al (1996) Nomenclature: vertebrate mediators of TGFβ family signals. Cell 87:173
Shi Y, Hata A, Lo RS, Massague J, Pavletich NP (1997) A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature 388:87
Lin LH, Chang KW, Cheng HW, Liu CJ (2019) Somatic SMAD4 mutations in head and neck carcinoma are associated with tumor progression. Front Oncol 9:1379
Hernanda PY, Chen K, Das AM et al (2015) SMAD4 exerts a tumor-promoting role in hepatocellular carcinoma. Oncogene 34:5055–5068
**a X, Wu W, Huang C et al (2015) SMAD4 and its role in pancreatic cancer. Tumor Biol 36:111–119
Salovaara R, Roth S, Loukola A et al (2002) Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut 51:56–59
Qin B, Lam SS, Lin K (1999) Crystal structure of a transcriptionally active Smad4 fragment. Structure 7:1493–1503
Maurice D, Pierreux CE, Howell M, Wilentz RE, Owen MJ, Hill CS (2001) Loss of Smad4 Function in pancreatic tumors C-terminal Truncation leads to decreased stability. J Biol Chem 276:43175–43181
Wardell CP, Fujita M, Yamada T et al (2018) Genomic characterization of biliary tract cancers identifies driver genes and predisposing mutations. J Hepatol 68:959–969
Weinberg BA, **u J, Lindberg MR et al (2019) Molecular profiling of biliary cancers reveals distinct molecular alterations and potential therapeutic targets. J Gastrointest Oncol 10:652
Zehir A, Benayed R, Shah RH et al (2017) Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature Med 23:703
Kumar R, Kumar R, Tanwar P et al (2020) Deciphering the impact of missense mutations on structure and dynamics of SMAD4 protein involved in pathogenesis of gall bladder cancer. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1740789
Rose PW, Beran B, Bi C et al (2011) The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res 39:D392–D401
Huang B (2009) MetaPocket: a meta approach to improve protein ligand binding site prediction. OMICS 13:325–330
Yang J, Roy A, Zhang Y (2012) BioLiP: a semi-manually curated database for biologically relevant ligand–protein interactions. Nucleic Acids Res 41:D1096–D1103
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33:W363–W367
Mozhayskiy V, Tagkopoulos I (2012) Horizontal gene transfer dynamics and distribution of fitness effects during microbial in silico evolution. BMC Bioinform 13:S13
Mukherjee G, Jayaram B (2013) A rapid identification of hit molecules for target proteins via physico-chemical descriptors. Phys Chem Chem Phys 15:9107–9116
Irwin JJ, Shoichet BK (2005) ZINC− a free database of commercially available compounds for virtual screening. J Chem Inf Mod 45:177–182
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Lipinski CA (2004) Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1:337–341
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 25:1656–1676
Schuttelkopf AW, Van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363
Hess B, Bekker H, Berendsen HJ, Fraaije JG (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Kumari R, Kumar R, Open Source Drug Discovery Consortium & Lynn A (2014) g_mmpbsa: A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54:1951–1962
Kumar R, Maurya R, Saran S (2019) Introducing a simple model system for binding studies of known and novel inhibitors of AMPK: a therapeutic target for prostate cancer. J Biomol Struct Dyn 37:781–795
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134
Sarshekeh AM, Advani S, Overman MJ et al (2017) Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS ONE 12:e0178275
Kumar R, Maurya R, Saran S (2017) Identification of novel inhibitors of the translationally controlled tumor protein (TCTP): insights from molecular dynamics. Mol BioSyst 13:510–524
Simmerling C, Strockbine B, Roitberg AE (2002) All-atom structure prediction and folding simulations of a stable protein. J Am Chem Soc 124:11258–11259
Dong YW, Liao ML, Meng XL, Somero GN (2018) Structural flexibility and protein adaptation to temperature: molecular dynamics analysis of malate dehydrogenases of marine molluscs. Proc Natl Acad Sci USA 115:1274–1279
Durrant JD, McCammon JA (2011) Molecular dynamics simulations and drug discovery. BMC Biol 9:71
Zhao H, Caflisch A (2015) Molecular dynamics in drug design. Eur J Med Chem 91:4–14
Meng XY, Zhang HX, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7:146–157
Chen YC (2015) Beware of docking! Trends Pharmacol Sci 36:78–95
Kumar R, Saran S (2018) Structure, molecular dynamics simulation, and docking studies of Dictyostelium discoideum and human STRAPs. J Cell Biochem 119:7177–7191
Wang C, Greene DA, **ao L, Qi R, Luo R (2018) Recent developments and applications of the MMPBSA method. Front Mol Biosci 4:87
McNicholas S, Potterton E, Wilson KS, Noble MEM (2011) Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallog 67:386–394
Gagan J, Van Allen EM (2015) Next-generation sequencing to guide cancer therapy. Genome Med 7:80
Sicklick JK, Fanta PT, Shimabukuro K, Kurzrock R (2016) Genomics of gallbladder cancer: the case for biomarker-driven clinical trial design. Cancer Metastasis Rev 35:263–275
Schror K (2011) Pharmacology and cellular/molecular mechanisms of action of aspirin and non-aspirin NSAIDs in colorectal cancer. Best Pract Res Clin Gastroenterol 25:473–484
Acknowledgement
Rakesh and Rahul thank Indian Council of Medical Research and University Grant Commission, respectively, for their research fellowships.
Funding
Indian Council of Medical Research, Grant/Award Number: 5/13/1/TF/2015/NCD-III.
Author information
Authors and Affiliations
Contributions
RK. and PT. conceptualized and designed the study. RK. and RK. performed the experiments. RK. analysed the data and wrote manuscript. PT. provided laboratory infrastructure. All authors read and approved the final version of manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Research involving human participants or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, R., Kumar, R. & Tanwar, P. Structural based screening of potential inhibitors of SMAD4: a step towards personalized medicine for gall bladder and other associated cancers. Mol Divers 25, 1945–1961 (2021). https://doi.org/10.1007/s11030-021-10210-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10210-w