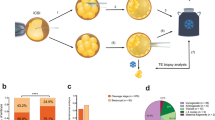
Abstract
Purpose
Providing feasible preimplantation genetic testing strategies for monogenic disorders (PGT-M) for prevention and control of genetic cancers.
Methods
Inclusion of families with a specific pathogenic mutation or a clear family history of genetic cancers. Identification of the distribution of hereditary cancer-related mutations in families through genetic testing. After a series of assisted reproductive measures such as down-regulation, stimulation, egg retrieval, and in vitro fertilization, a biopsy of trophectoderm cells from a blastocyst was performed for single-cell level whole-genome amplification (WGA). Then, the detection of chromosomal aneuploidies was performed by karyomap**. Construction of a haplotype-based linkage analysis to determine whether the embryo carries the mutation. Meanwhile, we performed CNV testing. Finally, embryos can be selected for transfer, and the results will be verified in 18–22 weeks after pregnancy.
Results
Six couples with a total of 7 cycles were included in our study. Except for cycle 1 of case 5 which did not result in a transferable embryo, the remaining 6 cycles produced transferable embryos and had a successful pregnancy. Four couples have had amniotic fluid tests to confirm that the fetus does not carry the mutation, while 1 couple was not tested due to insufficient pregnancy weeks. And the remaining couples had to induce labor due to fetal megacystis during pregnancy.
Conclusion
Our strategy has been proven to be feasible. It can effectively prevent transmission of hereditary cancer-related mutations to offspring during the prenatal stage.




Similar content being viewed by others
Data availability
The data that support the results of this study can be obtained from the corresponding author upon reasonable request.
References
Siegel RL, et al. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33.
Fitzmaurice C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the global burden of disease study. JAMA Oncol. 2018;4(11):1553–68.
Nagy R, Sweet K, Eng C. Highly penetrant hereditary cancer syndromes. Oncogene. 2004;23(38):6445–70.
Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer – analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85.
Garber JE, Offit K. Hereditary cancer predisposition syndromes. J Clin Oncol. 2005;23(2):276–92.
Fillon M. Breast cancer recurrence risk can remain for 10 to 32 years. CA Cancer J Clin. 2022;72(3):197–9.
Sakorafas GH, Tsiotou AG. Genetic predisposition to breast cancer: a surgical perspective. Br J Surg. 2000;87(2):149–62.
Handyside AH, et al. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature. 1990;344(6268):768–70.
Bickerstaff H, et al. Clinical application of preimplantation genetic diagnosis. Hum Fertil (Camb). 2001;4(1):24–30.
Treff NR, et al. Evaluation of targeted next-generation sequencing-based preimplantation genetic diagnosis of monogenic disease. Fertil Steril. 2013;99(5):1377–1384.e6.
Qubbaj W, et al. First successful application of preimplantation genetic diagnosis and haploty** for congenital hyperinsulinism. Reprod Biomed Online. 2011;22(1):72–9.
Wang Y, et al. A strategy using SNP linkage analysis for monogenic diseases PGD combined with HLA ty**. Clin Genet. 2020;98(2):138–46.
Yan L, et al. Live births after simultaneous avoidance of monogenic diseases and chromosome abnormality by next-generation sequencing with linkage analyses. Proc Natl Acad Sci U S A. 2015;112(52):15964–9.
Thornhill AR, et al. Karyomap** – a comprehensive means of simultaneous monogenic and cytogenetic PGD: comparison with standard approaches in real time for Marfan syndrome. J Assist Reprod Genet. 2015;32(3):347–56.
Harton GL, et al. ESHRE PGD consortium best practice guidelines for fluorescence in situ hybridization-based PGD. Hum Reprod. 2011;26(1):25–32.
Ricciardiello L, Ahnen DJ, Lynch PM. Chemoprevention of hereditary colon cancers: time for new strategies. Nat Rev Gastroenterol Hepatol. 2016;13(6):352–61.
Grunfeld E, et al. Family caregiver burden: results of a longitudinal study of breast cancer patients and their principal caregivers. Cmaj. 2004;170(12):1795–801.
Mishanina E, et al. Use of labour induction and risk of cesarean delivery: a systematic review and meta-analysis. Cmaj. 2014;186(9):665–73.
Wang Q, et al. Preimplantation genetic diagnosis using combined strategies on a breast cancer patient with a novel genomic deletion in BRCA2. J Assist Reprod Genet. 2014;31(12):1719–26.
Jasper MJ, Liebelt J, Hussey ND. Preimplantation genetic diagnosis for BRCA1 exon 13 duplication mutation using linked polymorphic markers resulting in a live birth. Prenat Diagn. 2008;28(4):292–8.
Rechitsky S, et al. Preimplantation genetic diagnosis for cancer predisposition. Reprod Biomed Online. 2002;5(2):148–55.
Girardet A, et al. First preimplantation genetic diagnosis of hereditary retinoblastoma using informative microsatellite markers. Mol Hum Reprod. 2003;9(2):111–6.
Shenfield F, et al. Taskforce 5: preimplantation genetic diagnosis. Hum Reprod. 2003;18(3):649–51.
Biesecker LG, Harrison SM. The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet Med. 2018;20(12):1687–8.
Cimadomo D, et al. The dawn of the future: 30 years from the first biopsy of a human embryo. The detailed history of an ongoing revolution. Hum Reprod Update. 2020;26(4):453–73.
Kokkali G, et al. Birth of a healthy infant following trophectoderm biopsy from blastocysts for PGD of beta-thalassaemia major. Hum Reprod. 2005;20(7):1855–9.
McArthur SJ, et al. Pregnancies and live births after trophectoderm biopsy and preimplantation genetic testing of human blastocysts. Fertil Steril. 2005;84(6):1628–36.
Carvalho F, et al. ESHRE PGT Consortium good practice recommendations for the detection of monogenic disorders. Hum Reprod Open. 2020;2020(3):hoaa018.
Antoniou A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–30.
Lee A, Moon BI, Kim TH. BRCA1/BRCA2 Pathogenic variant breast cancer: treatment and prevention strategies. Ann Lab Med. 2020;40(2):114–21.
Chen S, et al. Characterization of BRCA1 and BRCA2 mutations in a large United States sample. J Clin Oncol. 2006;24(6):863–71.
Narod SA. Which genes for hereditary breast cancer? N Engl J Med. 2021;384(5):471–3.
Carrera PM, Kantarjian HM, Blinder VS. The financial burden and distress of patients with cancer: understanding and step**-up action on the financial toxicity of cancer treatment. CA Cancer J Clin. 2018;68(2):153–65.
Bradley CJ. Cancer, financial burden, and medicare beneficiaries. J Clin Oncol. 2017;35(22):2461–2.
Li N, et al. Global burden of breast cancer and attributable risk factors in 195 countries and territories, from 1990 to 2017: results from the Global Burden of Disease Study 2017. J Hematol Oncol. 2019;12(1):140.
Chada AR, et al. Trends and outcomes for preimplantation genetic testing for monogenic disorders in the United States, 2014-2018. Fertil Steril. 2022;118(6):1190–3.
Lee I, et al. Utilization of preimplantation genetic testing for monogenic disorders. Fertil Steril. 2020;114(4):854–60.
Piyamongkol W, et al. Detailed investigation of factors influencing amplification efficiency and allele drop-out in single cell PCR: implications for preimplantation genetic diagnosis. Mol Hum Reprod. 2003;9(7):411–20.
Handyside AH, et al. Karyomap**: a universal method for genome wide analysis of genetic disease based on map** crossovers between parental haplotypes. J Med Genet. 2010;47(10):651–8.
Sherr CJ. Principles of tumor suppression. Cell. 2004;116(2):235–46.
Ethics of preimplantation genetic diagnosis for cancer. Lancet Oncol. 2006;7(8):611.
Luzzatto L, Pandolfi PP. Causality and chance in the development of cancer. N Engl J Med. 2015;373(1):84–8.
Acknowledgements
We would like to thank all the participants that took part in the study. This work was supported by the National Natural Science Foundation of China for the National Key R&D Program of China (2019YFA0110900) to Yingpu Sun.
Funding
This work was supported by funding from the National Natural Science Foundation of China for the National Key R&D Program of China (2019YFA0110900) to Y.-P.S.
Author information
Authors and Affiliations
Contributions
Chuanju Chen: conceptualization (supporting), data curation (lead), formal analysis (equal), validation (equal), visualization (lead), writing – original draft preparation (lead), writing – review and editing. Hao Shi: conceptualization (lead), formal analysis (equal), investigation (lead), methodology (lead), project administration (lead), supervision, writing – review and editing (lead). Wenbin Niu: formal analysis (equal), resources (equal). **ao Bao: formal analysis (equal), resources (equal). **gya Yang: validation (equal). Haixia **: formal analysis (equal), resources (equal). Wenyan Song: formal analysis (equal), resources (equal). Yingpu Sun: funding acquisition (lead), supervision (lead).
Corresponding author
Ethics declarations
Ethics approval
Ethical approval to conduct this retrospective study was obtained from the Internal Review Board of The First Affiliated Hospital of Zhengzhou University (Ethic no. 2023-KY-0361).
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
SUPPLEMENTAL Figure 1. Family trees of all family were included. (TIF 1252 kb)

ESM 2
SUPPLEMENTAL TABLE 1. The results of haplotype linkage analysis of case 1 by SNPs within 2M of upstream and downstream of the RET gene. (DOCX 30 kb)
ESM 3
SUPPLEMENTAL TABLE 2. The results of haplotype linkage analysis of case 1 by SNPs within 2M of upstream and downstream of the BRCA1 gene. (DOCX 21 kb)
ESM 4
SUPPLEMENTAL TABLE 3A. The results of haplotype linkage analysis of case 3 by SNPs within 2M of upstream and downstream of the RAD51D gene. SUPPLEMENTAL TABLE 3B. The results of haplotype linkage analysis of case 3 by SNPs within 2M of upstream and downstream of the TP53 gene. (DOCX 39 kb)
ESM 5
SUPPLEMENTAL TABLE 4. The results of haplotype linkage analysis of case 4 by SNPs within 2M of upstream and downstream of the RB1 gene. (DOCX 21 kb)
ESM 6
SUPPLEMENTAL TABLE 5A. The results of haplotype linkage analysis of case 5 by SNPs within 2M of upstream and downstream of the FH gene. SUPPLEMENTAL TABLE 5B. The results of haplotype linkage analysis of case 5 by SNPs within 1M of upstream and downstream of the NPHS1 gene from paternal chromosome. SUPPLEMENTAL TABLE 5C. The results of haplotype linkage analysis of case 5 by SNPs within 1M of upstream and downstream of the NPHS1 gene from maternal chromosome. (DOCX 38 kb)
ESM 7
SUPPMENTAL TABLE 6. The results of haplotype linkage analysis of case 6 by SNPs within 2M of upstream and downstream of the BRCA1 gene. (DOCX 28 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, C., Shi, H., Niu, W. et al. The preimplantation genetic testing for monogenic disorders strategy for blocking the transmission of hereditary cancers through haplotype linkage analysis by karyomap**. J Assist Reprod Genet 40, 2933–2943 (2023). https://doi.org/10.1007/s10815-023-02939-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-023-02939-0