Abstract
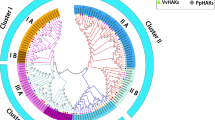
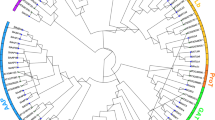
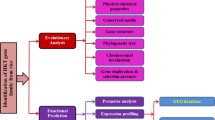
High-affinity K+ transporters (HKT) play an essential role in plant growth, development and response to abiotic stresses. Their main functions are to maintain the balance between Na+ and K+, and prevent sodium toxicity in root and leaf. Foxtail millet is a high stress-tolerant crop. It could reveal their potential functions in salt tolerance to investigation of the HKT genes. In this study, we identified 7 SiHKT genes in foxtail millet. These genes were classified into two subfamilies by phylogenetic analysis, prediction of conserved motif, and gene structure analysis. Comparative modeling and evaluation of HKT-transporters by AlphaFold2 informed further studies of the protein structure of SiHKTs. The gene expression of SiHKTs under salt stress showed that there were significant differences in SiHKTs in different tissues. At the early stage of salt stress, SiHKTs play a major role in transferring K+ into aboveground; and under salt stress condition, SiHKTs play an important part in preventing sodium toxicity in root. It implies that SiHKTs members might perform multiple functions. In addition, we predicted the transcription factors that regulate the gene expression of SiHKTs using WGCNA. This study provides the systematic description of the HKT gene family, which provides a framework for further functional analysis of SiHKTs, and is valuable for the development and utilization of the germplasm source of salt tolerance in foxtail millet.




Similar content being viewed by others
Data availability
The raw fastq data of RNA-seq of 11 xiaomi tissues were downloaded from the Bei**g Institute of Genomics Data Center (Accession numbers: CRA001953). The RNA-seq data of control and salt-stressed in foxtail millet were downloaded from NCBI SRA database (accession numbers PRJNA805389).
References
Al Nayef M, Solis C, Shabala L, Ogura T, Chen Z, Bose J, Maathuis FJM, Venkataraman G, Tanoi K, Yu M, Zhou M, Horie T, Shabala S (2020) Changes in Expression Level of OsHKT1;5 Alters Activity of Membrane Transporters Involved in K+ and Ca2+ Acquisition and Homeostasis in Salinized Rice Roots. Int J Mol Sci. https://doi.org/10.3390/ijms21144882
Ali A, Raddatz N, Aman R, Kim S, Park HC, Jan M, Baek D, Khan IU, Oh DH, Lee SY, Bressan RA, Lee KW, Maggio A, Pardo JM, Bohnert HJ, Yun DJ (2016) A single amino-acid substitution in the sodium transporter HKT1 associated with plant salt tolerance. Plant Physiol 171(3):2112–2126. https://doi.org/10.1104/pp.16.00569
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. https://doi.org/10.1093/nar/gkp335
Carpentier MC, Bousquet-Antonelli C, Merret R (2021) Fast and Efficient 5’P degradome library preparation for analysis of co-translational decay in Arabidopsis. Plants (basel). https://doi.org/10.3390/plants10030466
Cavrak VV, Lettner N, Jamge S, Kosarewicz A, Bayer LM, Mittelsten Scheid O (2014) How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet 10(1):e1004115. https://doi.org/10.1371/journal.pgen.1004115
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, **a R (2020) TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Chung BY, Simons C, Firth AE, Brown CM, Hellens RP (2006) Effect of 5’UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics 7:120. https://doi.org/10.1186/1471-2164-7-120
Cramer P (2021) AlphaFold2 and the future of structural biology. Nat Struct Mol Biol 28(9):704–705. https://doi.org/10.1038/s41594-021-00650-1
Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus A, Tester M (2007) The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 30(4):497–507. https://doi.org/10.1111/j.1365-3040.2007.01637.x
Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. Plant J 34(6):788–801. https://doi.org/10.1046/j.1365-313x.2003.01764.x
Hartmann L, Pedrotti L, Weiste C, Fekete A, Schierstaedt J, Göttler J, Kempa S, Krischke M, Dietrich K, Mueller MJ, Vicente-Carbajosa J, Hanson J, Dröge-Laser W (2015) Crosstalk between two bZIP signaling pathways orchestrates salt-induced metabolic reprogramming in Arabidopsis Roots. Plant Cell 27(8):2244–2260. https://doi.org/10.1105/tpc.15.00163
Hauser F, Horie T (2010) A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ 33(4):552–565. https://doi.org/10.1111/j.1365-3040.2009.02056.x
Huang L, Kuang L, Wu L, Shen Q, Han Y, Jiang L, Wu D, Zhang G (2020) The HKT transporter HvHKT1;5 negatively regulates salt tolerance. Plant Physiol 182(1):584–596. https://doi.org/10.1104/pp.19.00882
Janson G, Zhang C, Prado MG, Paiardini A (2017) PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics 33(3):444–446. https://doi.org/10.1093/bioinformatics/btw638
Julkowska MM, Klei K, Fokkens L, Haring MA, Schranz ME, Testerink C (2016) Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR-KISS gene. J Exp Bot 67(8):2127–2138. https://doi.org/10.1093/jxb/erw015
Kato N, Akai M, Zulkifli L, Matsuda N, Kato Y, Goshima S, Hazama A, Yamagami M, Guy HR, Uozumi N (2007) Role of positively charged amino acids in the M2D transmembrane helix of Ktr/Trk/HKT type cation transporters. Channels (austin) 1(3):161–171. https://doi.org/10.4161/chan.4374
Kole C (2021) Genomic designing for abiotic stress resistant cereal crops. Springer, Cham, pp 249–262. https://springer.longhoe.net/book/10.1007/978-3-030-75875-2
Li H, Xu G, Yang C, Yang L, Liang Z (2019) Genome-wide identification and expression analysis of HKT transcription factor under salt stress in nine plant species. Ecotoxicol Environ Saf 171:435–442. https://doi.org/10.1016/j.ecoenv.2019.01.008
Li M, Wang F, Ma J, Liu H, Ye H, Zhao P, Wang J (2022a) Comprehensive evolutionary analysis of CPP genes in Brassica napus L. and its two diploid progenitors revealing the potential molecular basis of allopolyploid adaptive advantage under salt stress. Front Plant Sci 13:873071. https://doi.org/10.3389/fpls.2022.873071
Li X, Gao J, Song J, Guo K, Hou S, Wang X, He Q, Zhang Y, Zhang Y, Yang Y, Tang J, Wang H, Persson S, Huang M, Xu L, Zhong L, Li D, Liu Y, Wu H, Diao X, Chen P, Wang X, Han Y (2022b) Multi-omics analyses of 398 foxtail millet accessions reveal genomic regions associated with domestication, metabolite traits, and anti-inflammatory effects. Mol Plant 15(8):1367–1383. https://doi.org/10.1016/j.molp.2022.07.003
Liu C, Chen K, Zhao X, Wang X, Shen C, Zhu Y, Dai M, Qiu X, Yang R, **ng D, Pang Y, Xu J (2019) Identification of genes for salt tolerance and yield-related traits in rice plants grown hydroponically and under saline field conditions by genome-wide association study. Rice (n y) 12(1):88. https://doi.org/10.1186/s12284-019-0349-z
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Mason MG, Jha D, Salt DE, Tester M, Hill K, Kieber JJ, Schaller GE (2010) Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in arabidopsis shoots. Plant J 64(5):753–763. https://doi.org/10.1111/j.1365-313X.2010.04366.x
Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A (2021) Pfam: The protein families database in 2021. Nucleic Acids Res 49(D1):D412–D419. https://doi.org/10.1093/nar/gkaa913
Mustafin ZS, Zamyatin VI, Konstantinov DK, Doroshkov AV, Lashin SA, Afonnikov DA (2019) Phylostratigraphic analysis shows the earliest origination of the abiotic stress associated genes in A. thaliana. Genes (basel). https://doi.org/10.3390/genes10120963
Pan Y, Michael TP, Hudson ME, Kay SA, Chory J, Schuler MA (2009) Cytochrome P450 monooxygenases as reporters for circadian-regulated pathways. Plant Physiol 150(2):858–878. https://doi.org/10.1104/pp.108.130757
Platten JD, Cotsaftis O, Berthomieu P, Bohnert H, Davenport RJ, Fairbairn DJ, Horie T, Leigh RA, Lin HX, Luan S, Mäser P, Pantoja O, Rodríguez-Navarro A, Schachtman DP, Schroeder JI, Sentenac H, Uozumi N, Véry AA, Zhu JK, Dennis ES, Tester M (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11(8):372–374. https://doi.org/10.1016/j.tplants.2006.06.001
Qin H, Wang J, Chen X, Wang F, Peng P, Zhou Y, Miao Y, Zhang Y, Gao Y, Qi Y, Zhou J, Huang R (2019) Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol 223(2):798–813. https://doi.org/10.1111/nph.15824
Riedelsberger J, Miller JK, Valdebenito-Maturana B, Piñeros MA, González W, Dreyer I (2021) Plant HKT channels: an updated view on structure, function and gene regulation. Int J Mol Sci. https://doi.org/10.3390/ijms22041892
Rubio F, Nieves-Cordones M, Horie T, Shabala S (2020) Doing “business as usual” comes with a cost: evaluating energy cost of maintaining plant intracellular K+ homeostasis under saline conditions. New Phytol 225(3):1097–1104. https://doi.org/10.1111/nph.15852
Sassi A, Mieulet D, Khan I, Moreau B, Gaillard I, Sentenac H, Véry AA (2012) The rice monovalent cation transporter OsHKT2;4: revisited ionic selectivity. Plant Physiol 160(1):498–510. https://doi.org/10.1104/pp.112.194936
Schachtman DP, Schroeder JI (1994) Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370(6491):655–658
Šimková K, Kim C, Gacek K, Baruah A, Laloi C, Apel K (2012) The chloroplast division mutant caa33 of Arabidopsis thaliana reveals the crucial impact of chloroplast homeostasis on stress acclimation and retrograde plastid-to-nucleus signaling. Plant J 69(4):701–712. https://doi.org/10.1111/j.1365-313X.2011.04825.x
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120
Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using clustalW and clustalX. Curr Protoc Bioinformatics. https://doi.org/10.1002/0471250953.bi0203s00
Tounsi S, Feki K, Saïdi MN, Maghrebi S, Brini F, Masmoudi K (2018) Promoter of the TmHKT1;4–A1 gene of Triticum monococcum directs stress inducible, developmental regulated and organ specific gene expression in transgenic Arbidopsis thaliana. World J Microbiol Biotechnol 34(7):99. https://doi.org/10.1007/s11274-018-2485-9
Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112:531–552. https://doi.org/10.1385/1-59259-584-7:531
**ao L, Shi Y, Wang R, Feng Y, Wang L, Zhang H, Shi X, **g G, Deng P, Song T, **g W, Zhang W (2022) The transcription factor OsMYBc and an E3 ligase regulate expression of a K+ transporter during salt stress. Plant Physiol. https://doi.org/10.1093/plphys/kiac283
Xu C, Luo M, Sun X, Yan J, Shi H, Yan H, Yan R, Wang S, Tang W, Zhou Y, Wang C, Xu Z, Chen J, Ma Y, Jiang Q, Chen M, Sun D (2022) SiMYB19 from Foxtail Millet (Setaria italica) confers transgenic rice tolerance to high salt stress in the field. Int J Mol Sci. https://doi.org/10.3390/ijms23020756
Yamaguchi T, Aharon GS, Sottosanto JB, Blumwald E (2005) Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc Natl Acad Sci USA 102(44):16107–16112. https://doi.org/10.1073/pnas.0504437102
Yang Z, Zhang H, Li X, Shen H, Gao J, Hou S, Zhang B, Mayes S, Bennett M, Ma J, Wu C, Sui Y, Han Y, Wang X (2020) A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nature Plants 6(9):1167–1178. https://doi.org/10.1038/s41477-020-0747-7
Yuan J, Shen C, Chen B, Shen A, Li X (2021) Genome-Wide Characterization and Expression Analysis of CAMTA Gene Family Under Salt Stress in Cucurbita moschata and Cucurbita maxima. Front Genet 12:647339. https://doi.org/10.3389/fgene.2021.647339
Zhang S, Tong Y, Li Y, Cheng ZM, Zhong Y (2019) Genome-wide identification of the HKT genes in five Rosaceae species and expression analysis of HKT genes in response to salt-stress in Fragaria vesca. Genes Genomics 41(3):325–336. https://doi.org/10.1007/s13258-018-0767-0
Funding
This research was funded by the National Natural Science Foundation of China (NSFC) (32001608), the Youth Fund Project on Application of Basic Research Project of Shanxi Province (Grant No. 201901D211362), and the Excellent doctors come to Shanxi to reward scientific research projects (Grant No. SXBYKY2022059).
Author information
Authors and Affiliations
Contributions
Experiments were designed by XL. Experiments were performed by YY, JC, HH and YZ. YY, RS and YL analyzed the data. YY wrote the manuscript. XL and HZ revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Communicated by Zhen Liang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, Y., Cheng, J., Han, H. et al. Genome-wide identification of the HKT transcription factor family and their response to salt stress in foxtail millet (Setaria italica). Plant Growth Regul 99, 113–123 (2023). https://doi.org/10.1007/s10725-022-00903-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-022-00903-z