Abstract
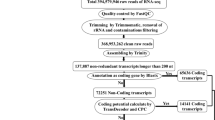
Long non-coding RNAs (lncRNAs) are a class of RNA regulatory molecules having roles in wide range of biological processes. They have been demonstrated to regulate gene expression at the posttranscriptional and transcriptional levels and to function in stress responses in plants and animals, but nothing is known about lncRNAs in Camelina (Camelina sativa L.), an emerging oil crop. Here, we report the first prediction of lncRNAs in the Camelina genome using comprehensive genomic approaches. We examined a Camelina drought stress cDNA library, and 5390 candidate Camelina sativa lncRNAs (CsalncRNAs) were identified, including 670 sense, 692 antisense, 1347 intergenic, and 2681 intronic harboring CsalncRNAs. The identified CsalncRNAs had an average nucleotide (nt) length of 497 bp and were mapped on each chromosome of C. sativa. Functional characterization through gene ontology (GO) and GO motif (GOMo) analysis of neighboring protein coding (PC) genes and motifs in the intergenic CsalncRNAs, respectively, indicated that these CsalncRNAs were involved in transcription-related activity, proteins, DNA and RNA binding, and abiotic/biotic stress response. Approximately 4.6% of CsalncRNA sequences were masked as repeat elements enriched with many repetitive sequences of transposable elements (TE), indicating the involvement of transposon silencing. Additionally, 55 intergenic CsalncRNAs were predicted as targets of miRNA, whereas nine target mimics were identified. Expression profiling of seven randomly selected CsalncRNAs using real-time quantitative polymerase chain reaction (RT-qPCR) showed tissue-specific expression, and these were highly up-regulated in Camelina leaves under extreme drought. Results of expression profiling indicated that these CsalncRNAs are involved in the progression of Camelina growth and development as well as its response to drought stress. Our results provide a basis for the functional study of lncRNAs in C. sativa that will serve as a valuable resource for future studies of the regulatory mechanisms underlying its growth and development as well as its stress response.






Similar content being viewed by others
References
Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M (2014) Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell 55(3):383–396
Bhattacharjee A, Khurana JP, Jain M (2016) Characterization of rice homeobox genes, OsHOX22 and OsHOX24, and over-expression of OsHOX24 in transgenic arabidopsis suggest their role in abiotic stress response. Front Plant Sci 7:627
Blackwell BJ, Lopez MF, Wang J, Krastins B, Sarracino D, Tollervey JR, Dobke M, Jordan IK, Lunyak VV (2012) Protein interactions with piALU RNA indicates putative participation of retroRNA in the cell cycle, DNA repair and chromatin assembly. Mob Genet Elements 2(1):26–35
Brosnan CA, Voinnet O (2009) The long and the short of noncoding RNAs. Curr Opin Cell Biol 21(3):416–425
Charon C, Moreno AB, Bardou F, Crespi M (2010) Non-protein-coding RNAs and their interacting RNA-binding proteins in the plant cell nucleus. Mol Plant 3(4):729–739
Chen B, Zhang Y, ZHang X, Jia S, Chen S, Kang L (2016) Genome-wide identification and developmental expression profiling of long noncoding RNAs during Drosophila metamorphosis. Sci Rep 6:23330. https://doi.org/10.1038/srep23330
Chung PJ, Jung H, Jeong DH, Ha SH, Choi YD, Kim JK (2016) Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genom 17:563
Collins-Silva JE, Lu C, Cahoon EB (2011) Camelina: a designer biotech oilseed crop. Inform 22:610–613
Dahro B, Wang F, Peng T, Liu J-H (2016) PtrA/NINV, an alkaline/neutral invertase gene of Poncirus trifoliata, confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency. BMC Plant Biol 16:76
Dai X, Zhao PX (2011) psRNAtarget: a plant small RNA target analysis server. Nucleic Acids Res 39:W155–W159
Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, **ao J, Zhang Q (2012) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid Rice. Proc Natl Acad Sci 109(7):2654–2659
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95(25):14863–14868
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio- Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39(8):1033–1037
Gong C, Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470(7333):284–288
Goodier JL, Cheung LE, Kazazian HH Jr (2013) Map** the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res 41(15):7401–7419
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458(7235):223–227
Huang W, Long L, Khatib H (2012) Genome-wide identification and initial characterization of bovine long non-coding RNAs from EST data. Anim Genet 43(6):674–682
Hutcheon C, Ditt RF, Beilstein M, Comai L, Schroeder J, Gold stein E, Shewmaker CK, Nguyen T, De Rocher J, Kiser J (2010) Polyploid genome of Camelina sativa revealed by isolation of fatty acid synthesis genes. BMC Plant Biol 10:233
Huttenhofer A, Schattner P, Polacek N (2005) Non-coding RNAs: hope or hype? Trends Genet 21(5):289–297
Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y (2013) A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25(10):4166–4182
Juan L, Wang G, Radovich M, Schneider BP, Clare SE, Wang Y, Liu Y (2013) Potential roles of microRNAs in regulating long intergenic noncoding RNAs. BMC Med Genet 6(Suppl 1):S7
Kagale S, Koh C, Nixon J, Bollina V, Clarke WE, Tuteja R, Spillane C, Robinson SJ, Links MG, Clarke C, Higgins Erin E, Huebert T, Sharpe AG, Parkin IAP (2014) The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat Commun 5:3706
Kagale S, Nixon J, Khedikar Y, Pasha A, Provart NJ, Clarke WE, Bollina V, Robinson SJ, Coutu C, Hegedus DD, Sharpe AG, Parkin IA (2016) The developmental transcriptome atlas of the biofuel crop Camelina sativa. Plant J 88(55):879–894
Kang C, Liu Z (2015) Global identification and analysis of long noncoding RNAs in diploid strawberry Fragaria vesca during flower and fruit development. BMC Genom 16:815
Kanth BK, Kumari S, Choi SH, Ha HJ, Lee GJ (2015) Generation and analysis of expressed sequence tags (ESTs) of Camelina sativa to mine drought stress-responsive genes. Biochem Biophys Res Commun 467(1):83–93
Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R et al (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316(5830):1484–1488
Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C (2013) Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9(4):e1003470
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12(4):656–664
Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30(5):814–822
Kim HS, Oh JM, Luan S, Carlson JE, Ahn SJ (2013) Cold stress causes rapid but differential changes in properties of plasma membrane H?-ATPase of camelina and rapeseed. J Plant Physiol 170(9):828–837
Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S et al (2013) Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152(3):570–583
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19(9):1639–1645
Kwak KJ, Kang H, Han KH, Ahn SJ (2013) Molecular cloning, characterization, and stress-responsive expression of genes encoding glycine-rich RNA-binding proteins in Camelina sativa L. Plant Physiol Biochem 68:44–51
Li J, Wu B, Xu J, Liu C (2014a) Genome-wide identification and characterization of long intergenic non-coding RNAs in Ganoderma lucidum. PLoS ONE 9(6):e99442
Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chetoor AM, Givan SA, Cole RA, Fowler JE, Evans MM, Scanlon MJ, Yu J, Schnable PS, Timmermans MC, Springer NM, Muehlbauer GJ (2014b) Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol 15(2):R40
Li S, Sun B, Li Y, Liu C, Wu X, Zhang D, Shi Y, Song Y, Buckler ES, Zhang Z, Wang T, Li Y (2016) Numerous genetic loci identified for drought tolerance in the maize nested association map** populations. BMC Genom 17(1):894
Li S, Yu X, Lei N, Cheng Z, Zhao P, He Y, Wang W, Peng M (2017) Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava. Sci Rep 7:45981. https://doi.org/10.1038/srep45981
Liu C, Muchhal US, Raghothama KG (1997) Differential expression of TPS11, a phosphate starvation-induced gene in tomato. Plant Mol Biol 33(5):867–874
Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24(11):4333–4345
Liu D, Wang L, Zhai H, Song X, He S, Liu Q (2014) A Novel a/b-hydrolase gene IbMas enhances salt tolerance in transgenic sweet potato. PLoS ONE 9(12):e115128
Liu X, Hao L, Li D, Zhu L, Hu S (2015) Long non-coding RNAs and their biological roles in plants. Genom Proteom Bioinform 13:137–147
Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, Zarnegar BJ, Boxer LD, Rios EJ, Tao S, Kretz M, Khavari PA (2015) A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell 32(6):693–706. https://doi.org/10.1016/j.devcel.2015.01.028
Lu X, Chen X, Mu M, Wang J, Wang X, Wang D, Yin Z, Fan W, Wang S, Guo L, Ye W (2016) Genome-wide analysis of long noncoding RNAs and their responses to drought stress in cotton (Gossypium hirsutum L.). PLoS ONE 11(6):e0156723
Luke B, Lingner J (2009) TERRA: telomeric repeat-containing RNA. EMBO J 28(17):2503–2510
Mehrotra S, Goyal V (2014) Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genom Proteom Bioinform 12(4):164–171
Mercer TR, Mattick JS (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20(3):300–307
Nonogaki H (2014) Seed dormancy and germination—emerging mechanisms and new hypotheses. Front Plant Sci 5:233
Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R (2010) Long non-coding RNAs with enhancer-like function in human. Cell 143(1):46–58
Ou LJ, Liu ZB, Zhang ZQ, Wei G, Zhang YP, Kang LY, Yang BZ, Yang S, Lv JH, Liu YH (2017) Noncoding and coding transcriptome analysis reveals the regulation roles of long noncoding RNAs in fruit development of hot pepper (Capsicum annuum L). Plant Growth Regul 83(1):141–156
Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM, Hatzigeorgiou AG (2013) DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res 41:D239–D245
Paul P, Dhandapani V, Choi SR, Lim YP (2016) Genome wide identification and functional prediction of long non-coding RNAs in Brassica rapa. Genes Genom 38(6):547–555
Pauli A, Rinn JL, Schier AF (2011) Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12(2):136–149
Pezer Z, Brajkovic J, Feliciello I, Ugarkovic D (2011) Transcription of satellite DNAs in insects. Prog Mol Subcell Biol 51:161–178
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136(4):629–641
Poudel S, Aryal N, Lu C (2015) Identification of MicroRNAs and transcript targets in Camelina sativa by deep sequencing and computational methods. PLoS ONE 10(3):e0121542
Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME (2015) lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res 43:D168–D173
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 8:145–166
Sana J, Faltejskova P, Svoboda M, Slaby O (2012) Novel classes of non-coding RNAs and cancer. J Transl Med 10:103
Shuai P, Liang D, Tang S, Zhang Z, Ye CY, Su Y, **a X, Yin W (2014) Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J Exp Bot 65(17):4975–4983
Song X, Sun L, Luo H, Ma Q, Zhao Y, Pei D (2016) Genome-wide identification and characterization of long non-coding RNAs from mulberry (Morus notabilis) RNA-seq Data. Genes (Basel) 7(3):11. https://doi.org/10.3390/genes7030011
Sousa C, Johansson C, Charon C, Manyani H, Sautter C, Kondorosi A (2001) Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol Cell Biol 21(1):354–366
Subburaj S, Kim AY, Lee S, Kim KN, Suh MC, Kim GJ, Lee GJ (2016) Identification of novel stress-induced microRNAs and their targets in Camelina sativa using computational approach. Plant Biotechnol Rep 10(3):155 – 169
Swiezewski S, Liu F, Magsin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462:799–802
Szcześniak MW, Rosikiewicz W, Makałowska I (2016) CANTATAdb: a collection of plant long non-coding RNAs. Plant Cell Physiol 57:e8. https://doi.org/10.1093/pcp/pcv201
Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang J, Lan F, Shi Y, Segal E, Chang H (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329(7274):689–693
Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R (2008) Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454(7200):126–130
Wang WC, Yang YW, Liu B, Sanyal A, Zimmerman RC, Chen Y, Lajoie BR, Protacia A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472(7341):120–126
Wang Y, Xu Z, Jiang J, Xu C, Kang J, **ao L, Wu M, **ong J, Guo X, Liu H (2013) Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell 25(1):69–80
Wang M, Yuan D, Tu L, Gao W, He Y, Hu H, Wang P, Liu N, Lindsey K, Zhang X (2015) Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.). New Phytol 207(4):1181–1197
Wang CY, Liu SR, Zhang XY, Ma YJ, Hu CG, Zhang JZ (2017) Genome-wide screening and characterization of long non-coding RNAs involved in flowering development of trifoliate orange (Poncirus trifoliata L. Raf.). Sci Rep 7:43226. https://doi.org/10.1038/srep43226
Wasaki J, Yonetani R, Shinano T, Kai M, Osaki M (2003) Expression of the OsPI1 gene, cloned from rice roots using cDNA microarray, rapidly responds to phosphorus status. New Phytol 158(2):239–248
Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlap** and adjacent genes. Cell 135(4):635–648
Wilusz JE, Sunwoo H, Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23(13):1494–1504
Wunderlich M, Groß-Hardt R, Schöffl F (2014) Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol Biol 85(6):541–550
**a H, Zhang L, Wu G, Fu C, Long Y, **ang J, Gan J, Zhou Y, Yu L, Li M (2016) Genome-wide identification and characterization of microRNAs and target genes in Lonicera japonica. PLoS ONE 11(10):e0164140
Yi X, Zhang Z, Ling Y, Xu W, Su Z (2015) PNRD: a plant noncoding RNA database. Nucleic Acids Res 43:D982–D989
Zaratiegui M, Irvin DV, Martienssen RA (2007) Noncoding RNAs and gene silencing. Cell 128(4):763–776
Zhang W, Han Z, Guo Q, Lin Y, Zheng Y, Wu F, ** W (2014a) Identification of maize long non-coding RNAs responsive to drought stress. PLoS ONE 9(6):e98958
Zhang Y, Liao J, Li Z, Yu Y, Zhang J, Li Q, Qu L, Shu W, Chen Y (2014b) Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol 15(12):512
Zhang H, Hu W, Hao J, Lv S, Wang C, Tong W, Wang Y, Wang Y, Liu X, Ji W (2016) Genome-wide identification and functional prediction of novel and fungi-responsive lincRNAs in Triticum aestivum. BMC Genom 17:238. https://doi.org/10.1186/s12864-016-2570-0
Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322(5902):750–756
Zou C, Wang Q, Lu C, Yang W, Zhang Y, Cheng H, Feng X, Prosper MA, Song G (2016) Transcriptome analysis reveals lng noncoding RNAs involved in fiber development in cotton (Gossypium arboreum). Sci China Life Sci 59(2):164–1671
Zubr J (1997) Oil-seed crop Camelina sativa. Ind Crops Prod 6(2):113–119
Zucchelli S, Fasolo F, Russo R, Cimatti L, Patrucco L, Takahashi H, Jones MH, Santoro C, Sblattero D, Cotella D, Persichetti F, Carninci P, Gustincich S (2015) SINEUPs are modular antisense long non-coding RNAs that increase synthesis of target proteins in cells. Front Cell Neurosci 9:174
Acknowledgements
This research was supported by the funds from Bio-industry Technology Development Program (No. 312033-5) and Golden Seed Project (Center for Vegetable Seed Development, No. 213003-05-1-SBW30), Ministry of Agriculture, Food and Rural Affairs (MAFRA) of Korea, and iPET (Korea Institute of Planning and Evaluation for Technology in Agriculture, Food and Rural Affairs).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10725_2018_410_MOESM1_ESM.tif
Supplementary Figure S1. Functional annotation of protein-coding genes identified as neighboring to the CsalncRNAs. All the genes are classified into three different categories as annotated by the GO terms. (A) Primary functions; (B) Biological processes; (C) Cellular components of the neighboring genes. (TIF 995 KB)
10725_2018_410_MOESM2_ESM.tif
Supplementary Figure S2. Hierarchical clustering analysis of all expressed CsalncRNAs and their nearest PC genes from the drought-induced Camelina leaf transcriptome library. Co-expression patterns of CsalncRNAs and their 5’ and 3’ neighboring PC genes were assessed by Log2 fold-change expression values from the EST transcriptome resource. In heatmap, red (A1–3) and green (B1–3) boxes represent CsalncRNA up-regulation: PC gene down-regulation and CsalncRNA down-regulation: PC gene up-regulation during drought conditions (10 kPa, 100 kPa, and Re-hyd), respectively. Reverse or positive correlation in expression between CsalncRNAs and neighboring PC genes are indicated by a blue bracket in heatmap. On the right-side panel, Venn analysis shows the number of CsalncRNAs that were up-regulated (A1–3) and down-regulated (B1–3) during each drought and in all drought conditions. Below, a color bar scale indicating either up- (red) or down- (green) regulation of expression. (TIF 2191 KB)
10725_2018_410_MOESM3_ESM.docx
Supplementary Table S1. List of primers designed and used in RT-qPCR analysis to determine the expression levels of lncRNAs in this study. (DOCX 12 KB)
10725_2018_410_MOESM5_ESM.xlsx
Supplementary Table S3. List of identified protein-coding genes detected immediately upstream and downstream of the CsalncRNAs. (XLSX 135 KB)
10725_2018_410_MOESM8_ESM.xlsx
Supplementary Table S6. List of functional encoding genes targeted by Camelina drought-responsive miRNAs regulating intergenic CsalncRNAs. (XLSX 48 KB)
Rights and permissions
About this article
Cite this article
Subburaj, S., Jeon, Y., Tu, L. et al. Genome-wide identification, functional prediction and expression profiling of long non-coding RNAs in Camelina sativa. Plant Growth Regul 86, 49–63 (2018). https://doi.org/10.1007/s10725-018-0410-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-018-0410-8