Abstract
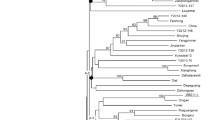
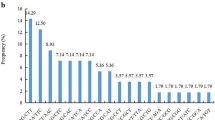
Taro [Colocasia esculenta (L.) Schott] is an old crop with high genetic diversity. However, the breeding of taro is limited by the lack of well-developed genetic markers. To help further research on taro germplasm resource management, we developed a set of SSR marker primers. Restriction site-associated DNA sequencing (RAD-seq) was used to identify SSRs from a taro variety. PCR and capillary electrophoresis were applied to evaluate the efficiency of the primers. A phylogenetic tree was constructed to analyse the evolutionary relationship of 30 taro varieties. A total of 4438 SSRs (simple sequence repeats)were identified using RAD sequencing. A total of 3919 pairs of primers were designed based on the SSR data. Trinucleotide repeats (49.1%) were the most abundant type of SSR, while AT was the most common motif type (327 instances). Among 50 randomly selected primer pairs, 46 primer pairs were able to amplify the corresponding bands. A phylogenetic tree cluster analysis using SSR markers mainly supported the classification of taro varieties according to their phenotypes and ecological distribution. The SSR markers identified in this study provide a useful resource for taro genetic and germplasm research and genetic breeding.


Similar content being viewed by others
References
Aggarwal RK, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V, Singh L (2006) Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor Appl Genet 114:359. https://doi.org/10.1007/s00122-006-0440-x
Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA (2016) Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet 17:81–92. https://doi.org/10.1038/nrg.2015.28
Barchi L, Lanteri S, Portis E, Acquadro A, Valè G, Toppino L, Rotino GL (2011) Identification of SNP and SSR markers in eggplant using RAD tag sequencing. BMC Genomics 12:304. https://doi.org/10.1186/1471-2164-12-304
Bonatelli IAS, Carstens BC, Moraes EM (2015) Using next generation RAD sequencing to isolate multispecies microsatellites for pilosocereus (Cactaceae). PLoS ONE 10:e0142602. https://doi.org/10.1371/journal.pone.0142602
Chaïr H, Traore RE, Duval MF, Rivallan R, Mukherjee A et al (2016) Genetic diversification and dispersal of taro (Colocasia esculenta (L.) Schott). PLoS ONE 11:e0157712. https://doi.org/10.1371/journal.pone.0157712
Chaudhary J, Alisha A, Bhatt V, Chandanshive S, Kumar N, Mir Z, Kumar A, Yadav SK, Shivaraj SM, Sonah H, Deshmukh R (2019) Mutation breeding in tomato: advances, applicability and challenges. Plants 8:128. https://doi.org/10.3390/plants8050128
Fang X, Huang K, Nie J, Zhang YL, Zhang YK, Li Y, Wang W, Xu X, Ruan R, Yuan X, Zhang Z, Yi Z (2019) Genome-wide mining, characterization, and development of microsatellite markers in Tartary buckwheat (Fagopyrum tataricum Garetn.). Euphytica 215:183. https://doi.org/10.1007/s10681-019-2502-6
Feng J, Zhao S, Li M, Zhang C, Qu H, Li Q, Li J, Lin Y, Pu Z (2020) Genome-wide genetic diversity detection and population structure analysis in sweetpotato (Ipomoea batatas) using RAD-seq. Genomics 112:1978–1987. https://doi.org/10.1016/j.ygeno.2019.11.010
Grimaldi IM, Muthukumaran S, Tozzi G, Nastasi A, Boivin N et al (2018) Literary evidence for taro in the ancient Mediterranean: a chronology of names and uses in a multilingual world. PLoS ONE 13(6):e0198333. https://doi.org/10.1371/journal.pone.0198333
Helmkampf M, Wolfgruber TK, Bellinger MR, Paudel R, Kantar MB, Miyasaka SC, Kimball HL, Brown A, Veillet A, Read A, Shintaku M (2018) Phylogenetic relationships, breeding implications, and cultivation history of Hawaiian Taro (Colocasia esculenta) through genome-wide SNP genoty**. J Hered 109:272–282. https://doi.org/10.1093/jhered/esx070
Hu KAN, Huang XF, Ke WD, Ding YI (2009) Characterization of 11 new microsatellite loci in taro (Colocasia esculenta). Mol Ecol Resour 9:582–584. https://doi.org/10.1111/j.1755-0998.2008.02441.x
Irwin SV, Kaufusi P, Banks K, de la Peña R, Cho JJ (1998) Molecular characterization of taro (Colocasia esculenta) using RAPD markers. Euphytica 99:183. https://doi.org/10.1023/A:1018309417762
Jansen van Rensburg WS, Shanahan P, Greyling M (2013) Molecular characterization of South African taro (Colocasia esculenta) germplasm using SSR markers. Acta Hortic 979:459–466. https://doi.org/10.17660/ActaHortic.2013.979.49
Jiang L, Wang L, Liu L, Zhu X, Zhai L, Gong Y (2012) Development and characterization of cDNA library based novel EST-SSR marker in radish (Raphanus sativus L.). Sci Hortic 140:164–172. https://doi.org/10.1016/j.scienta.2012.04.012
Kofler R, Schlotterer C, Lelley T (2007) SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23:1683–1685. https://doi.org/10.1093/bioinformatics/btm157
Kreike CM, Van Eck HJ, Lebot V (2004) Genetic diversity of taro, Colocasia esculenta (L.) Schott, in Southeast Asia and the Pacific. Theor Appl Genet 109:761–768. https://doi.org/10.1007/s00122-004-1691-z
Kumpatla SP, Mukhopadhyay S (2005) Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome 48:985–998. https://doi.org/10.1139/g05-060
Laval G, SanCristobal M, Chevalet C (2002) Measuring genetic distances between breeds: use of some distances in various short term evolution models. Genet Sel Evol 34:481–507. https://doi.org/10.1186/1297-9686-34-4-481
Lebot V, Aradhya KM (1991) Isozyme variation in taro (Colocasia esculenta (L.) Schott) from Asia and Oceania. Euphytica 56:55–66. https://doi.org/10.1007/BF00041744
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. https://doi.org/10.1093/bioinformatics/btl158
Liu S, An Y, Li F, Li S, Liu L, Zhou Q, Zhao S, Wei C (2018) Genome-wide identification of simple sequence repeats and development of polymorphic SSR markers for genetic studies in tea plant (Camellia sinensis). Mol Breed 38:59. https://doi.org/10.1007/s11032-018-0824-z
Loy TH, Spriggs M, Wickler S (1992) Direct evidence for human use of plants 28,000 years ago: starch residues on stone artefacts from the northern Solomon Islands. Antiquity 66:898–912. https://doi.org/10.1017/S0003598X00044811
Lu Z, Li W, Yang Y, Hu X (2011) Isolation and characterization of 19 new microsatellite loci in Colocasia esculenta (Araceae). Am J Bot 98:e239–e241. https://doi.org/10.3732/ajb.1100067
Luo R, Liu B, **e Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, **aoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18–18. https://doi.org/10.1186/2047-217X-1-18
Mace ES, Godwin ID (2002) Development and characterization of polymorphic microsatellite markers in taro (Colocasia esculenta). Genome 45:823–832. https://doi.org/10.1139/g02-045
Marriage TN, Hudman S, Mort ME, Orive ME, Shaw RG, Kelly JK (2009) Direct estimation of the mutation rate at dinucleotide microsatellite loci in Arabidopsis thaliana (Brassicaceae). Heredity 103:310–317. https://doi.org/10.1038/hdy.2009.67
Meger J, Ulaszewski B, Vendramin GG, Burczyk J (2019) Using reduced representation libraries sequencing methods to identify cpDNA polymorphisms in European beech (Fagus sylvatica L.). Tree Genet Genomes 15:7. https://doi.org/10.1007/s11295-018-1313-6
Miah G, Rafii MY, Ismail MR, Puteh AB, Rahim HA, Islam KN, Latif MA (2013) A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int J Mol Sci 14:22499–22528. https://doi.org/10.3390/ijms141122499
Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA (2007) Rapid and cost-effective polymorphism identification and genoty** using restriction site associated DNA (RAD) markers. Genome Res 17:240–248. https://doi.org/10.1101/gr.5681207
Onyango MG et al (2016) Genoty** of whole genome amplified reduced representation libraries reveals a cryptic population of Culicoides brevitarsis in the Northern Territory, Australia. BMC Genomics 17:769. https://doi.org/10.1186/s12864-016-3124-1
Parthiban S, Govindaraj P, Senthilkumar S (2018) Comparison of relative efficiency of genomic SSR and EST-SSR markers in estimating genetic diversity in sugarcane. 3 Biotech 8:144–144. https://doi.org/10.1007/s13205-018-1172-8
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. https://doi.org/10.1385/1-59259-192-2:365
Wang Z, Li J, Luo Z, Huang L, Chen X, Fang B, Li Y, Chen J, Zhang X (2011) Characterization and development of EST-derived SSR markers in cultivated sweetpotato (Ipomoea batatas). BMC Plant Biol 11:139. https://doi.org/10.1186/1471-2229-11-139
Xu J, Yang Y, Pu Y, Ayad WG, Eyzaguirre PB (2001) Genetic diversity in Taro (Colocasia esculenta Schott, Araceae) in China: an ethnobotanical and genetic approach. Econ Bot 55:14–31. https://doi.org/10.1007/BF02864543
Yagi M, Shirasawa K, Waki T, Kume T, Isobe S, Tanase K, Yamaguchi H (2017) Construction of an SSR and RAD marker-based genetic linkage map for carnation (Dianthus caryophyllus L.). Plant Mol Biol Rep 35:110–117. https://doi.org/10.1007/s11105-016-1010-2
You Y, Liu D, Liu H, Zheng X, Diao Y, Huang X, Hu Z (2015) Development and characterisation of EST-SSR markers by transcriptome sequencing in taro (Colocasia esculenta (L.) Schoot). Mol Breed 35:134. https://doi.org/10.1007/s11032-015-0307-4
Zeng S, **ao G, Guo J, Fei Z, Xu Y, Roe BA, Wang Y (2010) Development of a EST dataset and characterization of EST-SSRs in a traditional Chinese medicinal plant, Epimedium sagittatum (Sieb. Et Zucc.) Maxim. BMC Genomics 11:94. https://doi.org/10.1186/1471-2164-11-94
Zhao Z, Guo C, Sutharzan S, Li P, Echt CS, Zhang J, Liang C (2014) Genome-wide analysis of tandem repeats in plants and green algae. G3 (Bethesda) 4:67–78. https://doi.org/10.1534/g3.113.008524
Zheng XW, Cheng T, Yang L, Xu J, Tang J, **e K, Huang X, Bao Z, Zheng XF, Diao Y, You Y, Hu Z (2019) Genetic diversity and DNA fingerprints of three important aquatic vegetables by EST-SSR markers. Sci Rep 9:14074. https://doi.org/10.1038/s41598-019-50569-3
Acknowledgements
This study was supported by Natural Science Foundation of Guangxi Province (CN) (GuiTS201413)
Funding
This study was supported by Natural Science Foundation of Guangxi Province (CN) (GuiTS201413).
Author information
Authors and Affiliations
Contributions
WD and FH designed the experiments. WD, SW, ZQ, and QC performed the experiments. WD analysed the data. WD and FH wrote the paper. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical standard
Our manuscript is our original research work and academically satisfies all moral and ethical standards without plagiarizing or false data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dong, W., He, F., Wei, S. et al. Identification and characterization of SSR markers in taro [Colocasia esculenta (L.) Schott] by RAD sequencing. Genet Resour Crop Evol 68, 2897–2905 (2021). https://doi.org/10.1007/s10722-021-01162-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01162-z