Abstract
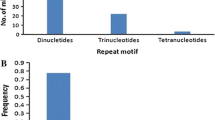
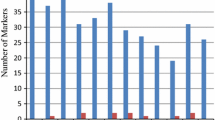
For breeding of salinity tolerant rice, halophytic species Oryza coarctata is considered as a valuable resource. Non-availability of molecular markers in the species is a major limitation and identification of markers applicable with wider gene pool will be resource-saving. Even after the recent advancement in high throughput genoty** techniques, sequence-tagged microsatellite sites (STMS) markers are considered as valuable resource for pre-breeding, especially when usable for a large number of species. Using a novel bioinformatics pipeline for cross transferable marker identification, whole genome sequences of nine Oryza species were surveyed with 23,499 STMS markers of rice. With this highly reproducible strategy, band sizes or polymorphism prediction among different species is possible before in-vitro validation. Only 359 STMS markers were cross-transferable to O. coarctata and 77 of those were common with O. sativa complex. These core markers were distributed over 11 chromosomes and nearly 76% were located within various genes of Oryza. The markers also showed unique genome specific polymorphism pattern with high levels of inter- and intraspecific variations in “AA” genome and complete absence of intra- or inter-population variations in O. coarctata (KKLL). The numbers of microsatellite motifs and the repeat numbers in different motifs were much lesser in O. coarctata leading to the exceptionally high level of polymorphism with all “AA” genome species. Two markers are also useful for species identification. The hyper-variable markers are effective resource for pre-breeding of O. sativa, O. glaberrima and new rice for Africa involving O. coarctata or any other species of O. sativa complex.







Similar content being viewed by others
References
Arbelaez JD, Moreno LT, Singh N, Tung CW, Maron LG, Ospina Y, Martinez CP, Grenier C, Lorieux M, McCouch S (2015) Development and GBS-genoty** of introgression lines (ILs) using two wild species of rice, O. meridionalis and O. rufipogon, in a common recurrent parent O. sativa cv. Curinga. Mol Breed 35:81. https://doi.org/10.1007/s11032-015-0276-7
Bal AR, Dutt SK (1986) Mechanism of salt tolerance in wild rice (Oryza coarctata Roxb.). Plant Soil 92:399–404. https://doi.org/10.1007/BF02372487
Balakrishnan D, Surapaneni M, Mesapogu S, Neelamraju S (2019) Development and use of chromosome segment substitution lines as a genetic resource for crop improvement. Theor Appl Genet 132:1–25. https://doi.org/10.1007/s00122-018-3219-y
Brar DS, Elloran RM, Talag JD, Abbasi F, Khush GS (1997) Cytogenetic and molecular characterization of an intergeneric hybrid between Oryza sativa L. and Porteresia coarctata (Roxb.) Tateoka. Rice Genet Newsl 14:43–44
Brar DS, Khush GS (2018) Wild relatives of rice: a valuable genetic resource for genomics and breeding research. In: Mondal TK, Henry RJ (eds) The wild Oryza genomes. Springer, pp 1–25
Brondani C, Rangel PH, Borba TC, Brondani RP (2003) Transferability of microsatellite and sequence tagged site markers in Oryza species. Hereditas 138:187–192. https://doi.org/10.1034/j.1601-5223.2003.01656.x
Chattopadhyay K, Nath D, Mohanta RL, Marndi BC, Singh DP, Singh ON (2015) Morpho-physiological and molecular variability in salt tolerant and susceptible popular cultivars and their derivatives at seedling stage and potential parental combinations in breeding for salt tolerance in rice. Cereal Res Commun 43:236–248. https://doi.org/10.1556/CRC.2014.0045
Chen J, Huang Q, Gao D, Wang J, Lang Y, Liu T, Li B, Bai Z, Goicoechea JL, Liang C, Chen C (2013) Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nature Commun 4:1595. https://doi.org/10.1038/ncomms2596
Chowrasia S, Rawal HC, Mazumder A, Gaikwad K, Sharma TR, Singh NK, Mondal TK (2018) Oryza coarctata Roxb. In: Mondal TK, Henry RJ (eds) The Wild Oryza Genomes. Springer, Cham, pp 87–104
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:39–40
Ganie SA, Karmakar J, Roychowdhury R, Mondal TK, Dey N (2016) An exploratory study on allelic diversity among rice and its wild species as well as relatives with simple sequence repeat and inter simple sequence repeat markers. Indian J Biotech 15:357–362. http://nopr.niscair.res.in/handle/123456789/39315
Ganie SA, Molla KA, Henry RJ, Bhat KV, Mondal TK (2019) Advances in understanding salt tolerance in rice. Theor Appl Genet 132:851–870. https://doi.org/10.1007/s00122-019-03301-8
Gao LZ, Zhang CH, Jia JZ (2005) Cross-species transferability of rice microsatellites in its wild relatives and the potential for conservation genetic studies. Genet Resour Crop Evol 52:931–940. https://doi.org/10.1007/s10722-003-6124-3
Gregorio GB, Senadhira D, Mendoza RD (1997) Screening rice for salinity tolerance. IRRI discussion paper series no. 22. International Rice Research Institute, Manila (Philippines), pp 1–30.
Islam T, Biswas S, Mita UH, Sarker RH, Rahman MS, Ali MA, Aziz KMS, Seraj ZI (2017) Characterization of progenies from intergeneric hybridization between Oryza sativa L. and Porteresia coarctata (Roxb.) Tateoka. Plant Tissue Cult Biotechnol 27:63–76. https://doi.org/10.3329/ptcb.v27i1.35013
Jacquemin J, Bhatia D, Singh K, Wing RA (2013) The International Oryza Map Alignment Project: development of a genus-wide comparative genomics platform to help solve the 9 billion people question. Curr Opin Plant Biol 16:147–156. https://doi.org/10.1016/j.pbi.2013.02.014
Jelodar N, Blackhall N, Hartman T, Brar D, Khush G, Davey M, Cocking E, Power J (1999) Intergeneric somatic hybrids of rice [Oryza sativa L. (+) Porteresia coarctata (Roxb.) Tateoka]. Theor Appl Genet 99:570–577. https://doi.org/10.1007/s001220051270
Jena KK (1994) Production of intergeneric hybrid between Oryza sativa L. and Porteresia coarctata T. Curr Sci 67:744–746. https://www.jstor.org/stable/24095851
Jiang SK, Huang C, Zhang XJ, Wang JY, Chen WF, Xu ZJ (2010) Development of a highly informative microsatellite (SSR) marker framework for rice (Oryza sativa L.) genoty**. Agric Sci China 9:1697–1704. https://doi.org/10.1016/S1671-2927(09)60268-6
** H, Tan G, Brar DS, Tang M, Li G, Zhu L, He G (2006) Molecular and cytogenetic characterization of an Oryza officinalis–O. sativa chromosome 4 addition line and its progenies. Plant Mol Biol 62:769–777. https://doi.org/10.1007/s11103-006-9056-4
Kellogg EA (2009) The evolutionary history of Ehrhartoideae, Oryzeae, and Oryza. Rice 2:1. https://doi.org/10.1007/s12284-009-9022-2
Kim H, Hurwitz B, Yu Y, Collura K, Gill N, SanMiguel P et al (2008) Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza. Genome Biol 9:R45. https://doi.org/10.1186/gb-2008-9-2-r45
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Li G, Hu W, Qin R, ** H, Tan G, Zhu L, He G (2008) Simple sequence repeat analyses of interspecific hybrids and MAALs of Oryza officinalis and Oryza sativa. Genetica 134:169–180. https://doi.org/10.1007/s10709-007-9222-x
Maclean J, Hardy B, Hettel G (2013) Rice Almanac: source book for one of the most important economic activities on earth, 3rd edn. CABI Publishing, UK
Malaviya DR, Roy AK, Kaushal P, Chakraborti M, Yadav A, Khare A, Dhir R, Khairnar D, George GP (2018) Interspecific compatibility barriers, development of interspecific hybrids through embryo rescue and lineage of Trifolium alexandrinum (Egyptian clover)- important tropical forage legume. Plant Breed 137:655–672. https://doi.org/10.1111/pbr.12616
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, ** of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207. https://doi.org/10.1093/dnares/9.6.199
Melaku G, Zhang S, Haileselassie T (2018) Comparative evaluation of Rice SSR markers on different Oryza species. J Rice Res Dev 1:38–48
Molla KA, Azharudheen TM, Ray S, Sarkar S, Swain A, Chakraborti M, Vijayan J, Singh ON, Baig MJ, Mukherjee AK (2019) Novel biotic stress responsive candidate gene based SSR (cgSSR) markers from rice. Euphytica 215:17. https://doi.org/10.1007/s10681-018-2329-6
Molla KA, Debnath AB, Ganie SA, Mondal TK (2015) Identification and analysis of novel salt responsive candidate gene based SSRs (cgSSRs) from rice Oryza sativaL. BMC Plant Biol 15:122. https://doi.org/10.1186/s12870-015-0498-1
Mondal TK, Rawal HC, Chowrasia S, Varshney D, Panda AK, Mazumdar A, Kaur H, Gaikwad K, Sharma TR, Singh NK (2018) Draft genome sequence of first monocot-halophytic species Oryza coarctata reveals stress-specific genes. Sci Rep 8:13698. https://doi.org/10.1038/s41598-018-31518-y
Mukherjee R, Mukherjee A, Bandyopadhyay S, Mukherjee S, Sengupta S, Ray S, Majumder AL (2019) Selective manipulation of the inositol metabolic pathway for induction of salt-tolerance in indica rice variety. Sci Rep 9:5358. https://doi.org/10.1038/s41598-019-41809-7
Narain A, Kar MK, Kaliaperumal V, Sen P (2016) Development of monosomic alien addition lines from the wild rice (Oryza brachyantha A. Chev. et Roehr.) for introgression of yellow stem borer (Scirpophaga incertulas Walker.) resistance into cultivated rice (Oryza sativa L.). Euphytica 209:603–613. https://doi.org/10.1007/s10681-016-1633-2
Neelam K, Mahajan R, Gupta V, Bhatia D, Gill BK, Komal R, Lore JS, Mangat GS, Singh K (2020) High-resolution genetic map** of a novel bacterial blight resistance gene xa-45(t) identified from Oryza glaberrima and transferred to Oryza sativa. Theor Appl Genet 133:689–705. https://doi.org/10.1007/s00122-019-03501-2
Oliveros JC (2007) Venny. An interactive tool for comparing lists with Venn's diagrams. Publicly available at https://bioinfogp.cnb.csic.es/tools/venny/index.html
Orjuela J, Garavito A, Bouniol M, Arbelaez JD, Moreno L, Kimball J, Wilson G, Rami JF, Tohme J, McCouch SR, Lorieux M (2010) A universal core genetic map for rice. Theor Appl Genet 120:563–572. https://doi.org/10.1007/s00122-009-1176-1
Parida SK, Dalal V, Singh AK, Singh NK, Mohapatra T (2009) Genic non-coding microsatellites in the rice genome: characterization, marker design and use in assessing genetic and evolutionary relationships among domesticated groups. BMC Genomics 10:140. https://doi.org/10.1186/1471-2164-10-140
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Science Publishers, Enfield, Montpellier, pp 43–76
Prusty MR, Kim SR, Vinarao R, Entila F, Egdane J, Diaz MG, Jena KK (2018) Newly identified wild rice accessions conferring high salt tolerance might use a tissue tolerance mechanism in leaf. Front Plant Sci 9:417. https://doi.org/10.3389/fpls.2018.00417
Rangan L, Sankararamasubramanian HM, Radha R, Swaminathan MS (2002) Genetic relationship of Porteresia coarctata Tateoka using molecular markers. Plant Biosyst 136:339–348
Ray S, Bose LK, Ray J, Ngangkham U, Katara JL, Samantaray S, Behera L, Anumalla M, Singh ON, Chen M, Wing RA, Mohapatra T (2016) Development and validation of cross-transferable and polymorphic DNA markers for detecting alien genome introgression in Oryza sativa. Mol Genet Genomics 291:1783–1794. https://doi.org/10.1007/s00438-016-1214-z
Rychlik WJ, Spencer WJ, Rhoads RE (1990) Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res 18:6409–6412. https://doi.org/10.1093/nar/18.21.6409
Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, Wakimoto H, Yang CC, Iwamoto M, Abe T, Yamada Y (2013) Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol 54:e6. https://doi.org/10.1093/pcp/pcs183
Sarkar S, Singh AM, Chakraborti M, Singh SK, Ahlawat AK, Singh GP (2014) Analysis of genetic diversity among the Indian bread wheat cultivars using microsatellite (SSR) markers. Indian J Genet Pl Br 74:502–505. https://doi.org/10.5958/0975-6906.2014.00877.3
Sayers EW, Agarwala R, Bolton EE, Brister JR, Canese K, Clark K, Connor R, Fiorini N, Funk K, Hefferon T, Holmes JB, Kim S, Kimchi A, Kitts PA, Lathrop S, Lu Z, Madden TL, Marchler-Bauer A, Phan L, Schneider VA, Schoch CL, Pruitt KD, Ostell J (2019) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res, 47(Database issue):D23-D28.https://doi.org/10.1093/nar/gky1069
Sengupta S, Majumder AL (2010) Porteresia coarctata (Roxb.) Tateoka, a wild rice: a potential model for studying salt-stress biology in rice. Plant Cell Environ 33:526–542. https://doi.org/10.1111/j.1365-3040.2009.02054.x
Sengupta S, Patra B, Ray S, Majumder AL (2008) Inositol methyl transferase from a halophytic wild rice, Porteresia coarctata Roxb. (Tateoka): regulation of pinitol synthesis under abiotic stress. Plant Cell Environ 31:1442–1458. https://doi.org/10.1111/j.1365-3040.2008.01850.x
Seraj ZI, Faruque MO, Hossain KG, Sarker RH, Devi T, Islam AS (1996) Attempted hybridization between Oryza satival. and P. Coarctata T. Int Rice Res Notes 21:35
Singh H, Deshmukh RK, Singh A, Singh AK, Gaikwad K, Sharma TR, Mohapatra T, Singh NK (2010) Highly variable SSR markers suitable for rice genoty** using agarose gels. Mol Breed 25:359–364. https://doi.org/10.1007/s11032-009-9328-1
Stein JC, Yu Y, Copetti D et al (2018) Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turn over and innovation across the genus Oryza. Nat Genet 50:285–296. https://doi.org/10.1038/s41588-018-0040-0
Tateoka T (1963) Taxonomic studies of Oryza. III. Key to the species and their enumeration. Bot Mag Tokyo 76:165–173
Tello-Ruiz MK, Stein J, Wei S, et al. (2016) Gramene 2016: comparative plant genomics and pathway resources. Nucleic Acids Res 44(Database issue):D1133–D1140.https://doi.org/10.1093/nar/gkv1179
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452. https://doi.org/10.1101/gr.184001
Umakanta N, Dash S, Parida M, Samantaray S, Nongthombam D, Yadav MK, Kumar A, Chidambaranathan P, Katara JL, Patra BC, Bose LK (2019) The potentiality of rice microsatellite markers in assessment of cross-species transferability and genetic diversity of rice and its wild relatives. Three Biotech. https://doi.org/10.1007/s13205-019-1757-x
Wing RA, Ammiraju JSS, Luo M, Kim H, Yu Y, Kudrna D et al (2005) The Oryza Map Alignment Project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol Biol 59:53–62. https://doi.org/10.1007/s11103-004-6237-x
Yu J, Hu S, Wang J, Wong GK, Li S et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296(5565):79–92. https://doi.org/10.1126/science.1068037
Zhang Y, Zhang S, Liu H, Fu B, Li L, **e M, Song Y, Li X, Cai J, Wan W, Kui L (2015) Genome and comparative transcriptomics of African wild rice Oryza longistaminata provide insights into molecular mechanism of rhizomatousness and self-incompatibility. Mol Plant 8:1683–1686. https://doi.org/10.1016/j.molp.2015.08.006
Zhu T, Xu PZ, Liu JP, Peng S, Mo XC, Gao LZ (2014) Phylogenetic relationships and genome divergence among the AA-genome species of the genus Oryza as revealed by 53 nuclear genes and 16 intergenic regions. Mol Phylogenetics Evol 70:348–361. https://doi.org/10.1016/j.ympev.2013.10.008
Zuccolo A, Sebastian A, Talag J, Yu Y, Kim H, Collura K, Kudrna D, Wing RA (2007) Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol Biol 7:152. https://doi.org/10.1186/1471-2148-7-152
Acknowledgements
The authors acknowledge the support of International Rice Research Institute, Philippines and Punjab Agriculture University, India for providing the germplasm of seven species used in the study. We are grateful to the teams of International Oryza Map Alignment Project, International Rice Genome Sequencing Project, Bei**g Genomics Institute and the Gramene database for making the Oryza genomic resources and analysis tools accessible for advancing research on individual loci. The academic and administrative support from the Director General, ICAR and Director, ICAR-National Rice Research Institute, Cuttack to the entire team and the Vice Chancellor of Utkal University, Bhubaneswar to the first author is also gratefully acknowledged.
Funding
This study was financially supported by institutional funding for Project 1.3 (2017–20) of ICAR-National Rice Research Institute. The University Grants Commission, Government of India supported the first author through Rajiv Gandhi National Fellowship for her Doctoral studies and the paper will be a part of her Ph.D. thesis.
Author information
Authors and Affiliations
Contributions
DD carried out the in-vitro validation; MC conceptualized and designed the experiments, arranged resources, analysed data and prepared draft manuscript; TKM, SR, D and JV carried out the Bioinformatics analyses; MKK and CP supervised the work; KC and KRC conducted salinity screening; DRP, LKB, SKD, BCP and BCM collected, imported and maintaining the germplasm used in the study; SS and MB provided support in wet lab works and preparation of tables and figures; TKM, SR, KC, SS and KRC critically reviewed the data and manuscript; DD, MC, TKM and SR made equal contribution for the work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
The authors declare that the experiments conducted for this publication comply with the current laws of India.
Conflict of interest
The authors declared that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Dalai, D., Chakraborti, M., Mondal, T.K. et al. The core set of sequence-tagged microsatellite sites markers between halophytic wild rice Oryza coarctata and Oryza sativa complex. Euphytica 217, 57 (2021). https://doi.org/10.1007/s10681-021-02790-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-021-02790-3