Abstract
Background and Aims
Acute pancreatitis (AP) is one of the common acute abdominal diseases with complicated pathogenesis. The purpose of this study is to identify the differentially expressed genes (DEGs) in the pancreas and underlying mechanisms.
Methods
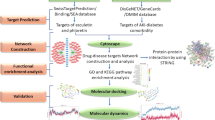
Gene expression profiles of GSE109227 and GSE65146 were available from GEO database. Then, an integrated analysis of these genes was performed, including gene ontology (GO) and KEGG pathway enrichment analysis, protein–protein interaction (PPI) network construction, core gene correlation analysis, transcription factors (TFs) prediction, and expression level evaluation in human organs.
Results
A total number of 92 differential expressed genes were screened from the datasets, including 81 up-regulated genes and 11 down-regulated genes. The up-regulated genes were mainly enriched in the biological process, such as sarcomere organization, actin cytoskeleton organization, tumor necrosis factor biosynthetic process, response to cytokine, cell–cell adhesion, and the cell migration, and also involved in some signaling pathways, including leukocyte transendothelial migration, proteoglycans in cancer, thyroid cancer, cell adhesion, tight junction, bladder cancer, amoebiasis, glycerolipid metabolism, and VEGF signaling pathway, while down-regulated genes were significantly enriched in the endoplasmic reticulum unfolded protein response, the oxidation–reduction, and no significant signaling pathways. CDH1 and CLDN4 were identified as core genes by PPI network analysis with MCODE plug-in, as well as GO and KEGG re-enrichment. For validation in Gene Expression Profiling Interactive Analysis (GEPIA), CDH1 and CLDN4 were interacting with each other and regulated by the predictive common TFs FOXP3 or USF2. The two core genes and USF2 were expressed in varied human organs including the pancreas, while FOXP3 was not detected in the normal human pancreatic tissues.
Conclusions
This study implied that core gene CDH1 and CLDN4, which might be regulated by FOXP3 or USF2, played a significant role in acute pancreatitis. They could be potential diagnostic and therapeutic targets for AP patients.





Similar content being viewed by others
References
Peery AF, Dellon ES, Lund J, et al. Burden of gastrointestinal disease in the United States: 2012 update. Gastroenterology. 2012;143:1.
Khurana A, Anchi P, Allawadhi P, et al. Yttrium oxide nanoparticles reduce the severity of acute pancreatitis caused by cerulein hyperstimulation. Nanomedicine. 2019;18:54–65.
Shen A, Kim H-J, Oh G-S, et al. Pharmacological stimulation of NQO1 decreases NADPH levels and ameliorates acute pancreatitis in mice. Cell death & disease. 2018;10:5.
Forsmark CE, Vege SS, Wilcox CM. Acute Pancreatitis. N Engl J Med. 2016;375:1972–1981.
Lankisch PG, Apte M, Banks PA. Acute pancreatitis. Lancet (London, England). 2015;386:85–96.
Garg PK, Singh VP. Organ Failure Due to Systemic Injury in Acute Pancreatitis. Gastroenterology. 2019;156:2008–2023.
Whitcomb DC. Genetic risk factors for pancreatic disorders. Gastroenterology. 2013;144:1292–1302.
Lowenfels AB, Lankisch PG, Maisonneuve P. What is the risk of biliary pancreatitis in patients with gallstones? Gastroenterology. 2000;119:879–880.
Lankisch PG, Lowenfels AB, Maisonneuve P. What is the risk of alcoholic pancreatitis in heavy drinkers? Pancreas. 2002;25:411–412.
Jancsó Z, Sahin-Tóth M. Mutation That Promotes Activation of Trypsinogen Increases Severity of Secretagogue-Induced Pancreatitis in Mice. Gastroenterology. 2019;158:1083–1094.
Saluja A, Dudeja V, Dawra R, Sah RP. Early Intra-Acinar Events in Pathogenesis of Pancreatitis. Gastroenterology. 2019;156:1979–1993.
Pelaez-Luna M, Robles-Diaz G, Canizales-Quinteros S, Tusié-Luna MT. PRSS1 and SPINK1 mutations in idiopathic chronic and recurrent acute pancreatitis. World J Gastroenterol. 2014;20:11788–11792.
Jalaly NY, Moran RA, Fargahi F, et al. An Evaluation of Factors Associated With Pathogenic PRSS1, SPINK1, CTFR, and/or CTRC Genetic Variants in Patients With Idiopathic Pancreatitis. Am J Gastroenterol. 2017;112:1320–1329.
Kujko AA, Berki DM, Oracz G, et al. A novel pSer282Pro variant is associated with autosomal dominant hereditary pancreatitis. Gut. 2017;66:1728–1730.
**ao Y, Yuan W, Yu B, et al. Targeted Gene Next-Generation Sequencing in Chinese Children with Chronic Pancreatitis and Acute Recurrent Pancreatitis. J Pediatr. 2017;191:1.
Upton GJG, Harrison AP. Motif effects in Affymetrix GeneChips seriously affect probe intensities. Nucleic Acids Res. 2012;40:9705–9716.
Fan L, Hui X, Mao Y, Zhou J. Identification of Acute Pancreatitis-Related Genes and Pathways by Integrated Bioinformatics Analysis. Dig Dis Sci. 2019;2019:1–13. https://doi.org/10.1007/s10620-019-05928-5
Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29.
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114.
Huang DW, Sherman BT, Tan Q, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–175.
Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815.
Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382.
Bandettini WP, Kellman P, Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson. 2012;14:83.
Capaldo CT, Farkas AE, Nusrat A. Epithelial adhesive junctions. F1000 Prime Rep. 2014;6:1.
Wheelock MJ, Johnson KR. Cadherins as modulators of cellular phenotype. Annu Rev Cell Dev Biol. 2003;19:207–235.
Wong SHM, Fang CM, Chuah L-H, Leong CO, Ngai SC. E-cadherin: its dysregulation in carcinogenesis and clinical implications. Crit Rev Oncol Hematol. 2018;121:11–22.
Braga V. Spatial integration of E-cadherin adhesion, signalling and the epithelial cytoskeleton. Curr Opin Cell Biol. 2016;42:138–145.
Nakada S, Tsuneyama K, Kato I, et al. Identification of candidate genes involved in endogenous protection mechanisms against acute pancreatitis in mice. Biochem Biophys Res Commun. 2010;391:1342–1347.
Serrill JD, Sander M, Shih HP. Pancreatic Exocrine Tissue Architecture and Integrity are Maintained by E-cadherin During Postnatal Development. Sci Rep. 2018;8:13451.
Mayerle J, Schnekenburger J, Krüger B, et al. Extracellular cleavage of E-cadherin by leukocyte elastase during acute experimental pancreatitis in rats. Gastroenterology. 2005;129:1251–1267.
Beeman N, Webb PG, Baumgartner HK. Occludin is required for apoptosis when claudin-claudin interactions are disrupted. Cell death & disease. 2012;3:e273.
Schaefer I-M, Agaimy A, Fletcher CD, Hornick JL. Claudin-4 expression distinguishes SWI/SNF complex-deficient undifferentiated carcinomas from sarcomas. Mod Pathol. 2017;30:539–548.
Lashhab R, Rumley AC, Arutyunov D, et al. The kidney anion exchanger 1 affects tight junction properties via claudin-4. Sci Rep. 2019;9:3099.
**a X-M, Wang F-Y, Wang Z-K, Wan H-J, Xu W-A, Lu H. Emodin enhances alveolar epithelial barrier function in rats with experimental acute pancreatitis. World J Gastroenterol. 2010;16:2994–3001.
Sonika U, Goswami P, Thakur B, et al. Mechanism of Increased Intestinal Permeability in Acute Pancreatitis: alteration in Tight Junction Proteins. J Clin Gastroenterol. 2017;51:461–466.
Wing JB, Tanaka A, Sakaguchi S. Human FOXP3 Regulatory T Cell Heterogeneity and Function in Autoimmunity and Cancer. Immunity. 2019;50:302–316.
Bluestone JA. FOXP3, the Transcription Factor at the Heart of the Rebirth of Immune Tolerance. J Immunol. 2017;198:979–980.
Kim J-H, Hwang J, Jung JH, Lee H-J, Lee DY, Kim S-H. Molecular networks of FOXP family: dual biologic functions, interplay with other molecules and clinical implications in cancer progression. Mol Cancer. 2019;18:180.
Kuwahara T, Hazama S, Suzuki N, et al. Correction: intratumoural-infiltrating CD4 + and FOXP3 + T cells as strong positive predictive markers for the prognosis of resectable colorectal cancer. British journal of cancer. 2019;121:983–984.
Ying L, Yan F, Meng Q, et al. PD-L1 expression is a prognostic factor in subgroups of gastric cancer patients stratified according to their levels of CD8 and FOXP3 immune markers. Oncoimmunology. 2018;7:e1433520.
Yang S, Liu Y, Li M-Y, et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 2017;16:124.
Funding
This work was supported by National Natural Science Foundation of China [Grant Nos. 81600501, 81670581]; Medical Guiding project of Science and Technology Commission of Shanghai Municipality [Grant number 16411970700, 18411966400]; and Key Discipline Construction Project of Shanghai Municipal Commission of Health and Family Planning [Grant Nos. 2016ZB0206].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ji, R., Chen, Y., Chen, W. et al. Identification of Significant Genes and Pathways in Acute Pancreatitis via Bioinformatical Analysis. Dig Dis Sci 66, 3045–3053 (2021). https://doi.org/10.1007/s10620-020-06598-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06598-4