Abstract
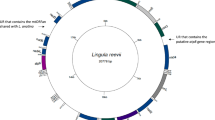
Brachyuran crabs comprise the most species-rich clades among extant Decapoda and are divided into several major superfamilies. However, the phylogeny of Brachyuran remains controversial, comprehensive analysis of the overall phylogeny is still lacking. Complete mitochondrial genome (mitogenome) can indicate phylogenetic relationships, as well as useful information for gene rearrangement mechanisms and molecular evolution. In this study, we firstly sequenced and annotated the complete mitogenome of Macrophthalmus abbreviatus (Brachyura; Macrophthalmidae). The mitogenome length of M. abbreviatus is 16,322 bp, containing the entire set of 37 genes and a control region typically observed in Brachyuran mitogenomes. The genome composition of M. abbreviatus was highly A+T biased 76.3% showing positive AT-skew (0.033) and negative GC-skew (− 0.351). In M. abbreviatus mitogenome, most tRNA genes were folded into the clover-leaf secondary structure except trnH, trnS1 and trnC, which was similar to the other species in Macrophthalmidae. Phylogenetic analysis showed that all families form a monophyletic, and Varunidae and Macrophthalmidae clustered into a monophyletic clade as sister groups. Comparative analyses of rearrangement among Brachyura revealed that Varunidae (Grapsoidea) and Macrophthalmidae (Ocypodoidea) had the same gene order, which reinforced the result of phylogeny. The combined results of two aspects revealed that the polyphyly of Ocypodoidea and Grapsoidea were well supported. In general, the results obtained in this research will contribute to further studies on molecular based for the classification and gene rearrangements of Macrophthalmidae or even Brachyura.






Similar content being viewed by others
References
Arndt A, Smith MJ (1998) Mitochondrial gene rearrangement in the sea cucumber genus Cucumaria. Mol Biol Evol 15:1009–1016
Altschul SF, Madden TL, Schäffer AA, Zhang JH, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein databases search programs. Nucleic Acids Res 25:3389–3402
Bai J, Xu S, Nie Z, Wang Y, Zhu C, Wang Y, Min W, Cai Y, Zou J, Zhou X (2018) The complete mitochondrial genome of Huananpotamon lichuanense (Decapoda: Brachyura) with phylogenetic implications for freshwater crabs. Gene 646:217–226
Basso A, Babbucci M, Pauletto M, Riginella E, Patarnello T, Negrisolo E (2017) The highly rearranged mitochondrial genomes of the crabs Maja crispata and Maja squinado (Majidae) and gene order evolution in Brachyura. Sci Rep 7:4096
Bernt M, Merkle D, Ramsch K, Fritzsch G, Perseke M, Bernhard D, Schlegel M, Stadler PF, Middendorf M (2007) CREx: inferring genomic rearrangements based on common intervals. Bioinformatics 23:2957–2958
Boudreau SA, Worm B (2012) Ecological role of large benthic decapods in marine ecosystems: a review. Mar Ecol Progr 469:195–213
Chen J, **ng Y, Yao W, Zhang C, Zhang Z, Jiang G, Ding Z (2018) Characterization of four new mitogenomes from Ocypodoidea & Grapsoidea, and phylomitogenomic insights into thoracotreme evolution. Gene 675:27–35
Chen Z, Liu YQ, Wu Y, Fan S, Li H (2020) Novel tRNA gene rearrangements in the mitochondrial genome of Camarochiloides weiweii (Hemiptera: Pachynomidae). Int J Biol Macromol 165:1738–1744
Conde J, Carmona-Suárez C (2005) The natural diet of Arenaeus Cribrarius (Decapoda, Brachyura, Portunidae) on two arid beaches in western Venezuela. Crustaceana 78:525–541
Cumberlidge N, Boyko CB, Harvey AW (2002) A new genus and species of freshwater crab (Crustacea, Decapoda, Potamoidea) from northern Madagascar, and a second new species associated with Pandanus leaf axils. Ann Mag Nat Hist 36:65–77
Diele K, Koch V (2010) Comparative population dynamics and life histories of north Brazilian Mangrove Crabs, Genera Uca and Ucides (Edgar, Robert C (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 5:1792–1797
Fenn JD, Cameron SL, Whiting MF (2010) The complete mitochondrial genome sequence of the Mormon cricket (Anabrus simplex: Tettigoniidae: Orthoptera) and an analysis of control region variability. Insect Mol Biol 16:239–252
Gissi C, Iannelli F, Pesole G (2008) Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 101:301–320
Guinot D (1977) Propositions pour une nouvelle classification des Crustace´s De´capodes Brachyoures. Comp Rend Acad Sci Paris D 285:1049–1052
Guinot D (1978) Principes d’une classification évolutive des Crustacés Decapodes Brachyoures. Bull Biol De La France Et De La Belg 112:211–292
Guinot D, Marcos T, Peter C (2013) Significance of the sexual openings and supplementary structures on the phylogeny of brachyuran crabs (Crustacea, Decapoda, Brachyura), with new nomina for higher-ranked podotreme taxa. Zootaxa 3665:1
Huelsenbeck JP (2012) Efficient Bayesian phylgenetic inference and model choice across a large model space. Syst. Biol. 61:539–542
Ki JS, Dahms HU, Hwang JS, Lee JS (2009) The complete mitogenome of the hydrothermal vent crab Xenograpsus testudinatus (Decapoda, Brachyura) and comparison with brachyuran crabs. Comp Biochem Physiol D Genomics Proteomics 4:290–299
Kong X, Dong X, Zhang Y, Shi W, Wang Z, Yu Z (2009) A novel rearrangement in the mitochondrial genome of tongue sole, Cynoglossus semilaevis: control region translocation and a tRNA gene inversion. Genome 52:975–984
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lavrov DV, Boore JL, Brown WM (2002) Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: duplication and nonrandom loss. Mol Biol Evol 19:163–169
Li L, Guo-Yue YU, Mcavoy TJ, Reardon RC, Yun WU, Salom SM, **g-Fu HE (2011) The biological characteristics, habitat and food selection of Tetraphleps galchanoides (Hemiptera: Anthocoridae). Acta Entomol Sin 54:800–808
Ma YK, Qin J, Lin TY, Ng PKL, Chu KH, Tsang LM (2019) Phylogenomic analyses of brachyuran crabs support early divergence of primary freshwater crabs. Mol Phylogenet Evol 135:62–66
María VL, Goldemberg AL, Alcira OD (2011) The claw closer muscle of Neohelice granulata (Grapsoidea, Varunidae): a morphological and histochemical study. Acta Zool 92:126–133
Mariappan P, Balasundaram CB (2000) Decapod crustacean chelipeds: an overview. J Biosci 25:301–313
Markert A, Raupach MJ, Segelken-Voigt A, Wehrmann A (2014) Molecular identification and morphological characteristics of native and invasive Asian brush-clawed crabs (Crustacea: Brachyura) from Japanese and German coasts: Hemigrapsus penicillatus (De Haan, 1835) versus Hemigrapsus takanoi Asakura & Watanabe 2005. Org Divers Evol 14:369–382
Matthias B, Alexander D, Frank J, Fabian E, Catherine F, Guido F, Joern P, Martin M, Peter FS (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol 69:313–319
Ming TL, Schubart CD, Ahyong ST, Lai JCY, Au EYC, Tin-Yam C, Ng PKL, Hou CK (2014) Evolutionary history of true crabs (Crustacea: Decapoda: Brachyura) and the origin of freshwater crabs. Mol Biol Evol 31:1173–1187
Ng PKL, Guinot D, Davie PJF (2008) Systema Brachyurorum: part I. An annotated checklist of extant brachyuran crabs of the world. Raffles B Zool 17:1–286
Okajima Y, Kumazawa Y (2010) Mitochondrial genomes of acrodont lizards: timing of gene rearrangements and phylogenetic and biogeographic implications. Bmc Evol Biol 10:141
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358
Rice AL (1980) Crab zoeal morphology and its bearing on the classification of the Brachyura. Trans Zool Soc Lond 35:271–372
Robert MJ, Schulte JA, Allan L, Ananjeva NB, Yuezhao W, Rohan P, Nasrullah RP, Papenfuss TJ (2000) Evaluating trans-tethys migration: an example using acrodont lizard phylogenetics. Syst Biol 49:233–256
Schubart CD, Cannicci S, Vannini M, Fratini S (2010) Molecular phylogeny of grapsoid crabs (Decapoda, Brachyura) and allies based on two mitochondrial genes and a proposal for refraining from current superfamily classification. J Zool Syst Evol Res 44:193–199
Schuwerack PM, Barnes RSK, Underwood GJC, Jones PW (2006) Gender and species differences in sentinel crabs (Macrophthalmus) feeding on an Indonesian mudflat. J Crustacean Biol 26:119–123
Shi W, Dong XL, Wang ZM, Miao XG, Kong XY (2013) Complete mitogenome sequences of four flatfishes (Pleuronectiformes) reveal a novel gene arrangement of L-strand coding genes. BMC Evol Biol 13:173
Shi W, Miao XG, Kong XY (2014) A novel model of double replications and random loss accounts for rearrangements in the Mitogenome of Samariscus latus (Teleostei: Pleuronectiformes). Bmc Genomics 15:352
Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT (2006) Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst 37:545–579
Spears T, Abele LG, Kim W (1992) The monophyly of brachyuran crabs: a phylogenetic study based on 18s rRNA. Syst Biol 41:446–461
Sternberg RV, Cumberlidge N (2001) Notes on the position of the true freshwater crabs within the brachyrhynchan Eubrachyura (Crustacea: Decapoda: Brachyura). Springer, Berlin, pp 21–39
Sternberg RV, Cumberlidge N, Rodriguez G (2010) On the marine sister groups of the freshwater crabs (Crustacea: Decapoda: Brachyura). J Zool Syst Evol Res. https://doi.org/10.1046/j.1439-0469.1999.95092.x
Sun HY, Kaiya Z, Daxiang S (2003) Mitochondrial genome and phylogenetic reconstruction of arthropods. Zool Res 24:467–479
Sun S, Hui M, Wang M, Sha Z (2018) The complete mitochondrial genome of the alvinocaridid shrimp Shinkaicaris leurokolos (Decapoda, Caridea): insight into the mitochondrial genetic basis of deep-sea hydrothermal vent adaptation in the shrimp. Comp Biochem Physiol D Genomics Proteomics 25:42–52
Takeda M, Miyake S (1970) Crabs from the East China Sea. IV Gymnopleura, Dromiacea and Oxystomata. J Fac Agric Kyushu Univ 16:193–235
Tan MH, Gan HM, Lee YP, Linton S, Austin CM (2018) ORDER within the chaos: insights into phylogenetic relationships within the Anomura (Crustacea: Decapoda) from mitochondrial sequences and gene order rearrangements. Mol Phylogenet Evol 127:320
Tsang LM, Schubart CD, Ahyong ST, Lai JCY, Au EYC, Chan T, Ng PKL, Chu KH (2014) Evolutionary history of true crabs (Crustacea: Decapoda: Brachyura) and the origin of freshwater crabs. Mol Biol Evol 31:1173–1187
Wang Z, Shi X, Tao Y, Wu Q, Bai Y, Guo H, Tang D (2018a) The complete mitochondrial genome of Parasesarma pictum (Brachyura: Grapsoidea: Sesarmidae) and comparison with other Brachyuran crabs. Genomics 111:799–807
Wang ZF, Wang ZQ, Shi XJ, Wu Q, Tao Y, Guo H, Ji CY, Bai YZ (2018b) Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura. Int J Biol Macromol 118:31–40
Wang Q, Tang D, Guo H, Wang J, Xu X, Wang Z (2020a) Comparative mitochondrial genomic analysis of Macrophthalmus pacificus and insights into the phylogeny of the Ocypodoidea & Grapsoidea. Genomics 112:82–91
Wang Z, Shi X, Guo H, Tang D, Bai Y, Wang Z (2020b) Characterization of the complete mitochondrial genome of Uca lacteus and comparison with other Brachyuran crabs. Genomics 112:10–19
**n ZZ, Liu Y, Zhang DZ, Chai XY, Wang ZF, Zhang HB, Zhou CL, Tang BP, Liu QN (2017) Complete mitochondrial genome of Clistocoeloma sinensis (Brachyura: Grapsoidea): gene rearrangements and higher-level phylogeny of the Brachyura. Sci Rep 7:4128
Yang ML, **gming XU, Bin WU, Hong** P (2014) A preliminary study on crabs diversity for Beibu Gulf Mangrove. Sichuan J Zool
Zhang Y, Bi Y, Feng M (2005) The complete mitochondrial genome of Lophosquillia costata (Malacostraca: Stomatopoda) from China and phylogeny of stomatopods. Mitochondrial DNA Part B 5:2495–2497
Zhang Y, Gong L, Lu X, Jiang L, Zhang X (2020) Gene rearrangements in the mitochondrial genome of Chiromantes eulimene (Brachyura: Sesarmidae) and phylogenetic implications for Brachyura. Int J Biol Macromol 162:704–714
Zhong S, Zhao Y, Zhang Q (2018) The first complete mitochondrial genome of Dorippoidea from Orithyia sinica (Decapoda: Orithyiidae). Mitochondrial DNA B 3:554–555
Acknowledgements
This study was funded by the National Natural Science Foundation of China (Grant Number 31702014), and Doctoral Scientific Research Foundation of Yancheng Teachers University to ZFW, and Open Foundation of Jiangsu Key Laboratory for Bioresources of Saline Soils (Grant Number JKLBS2019006).
Author information
Authors and Affiliations
Contributions
ZFW, XYX, QW and JW designed and conceived the experiment. ZFW, XYX, JYX and YJL performed the data analysis and draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no competing interest.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10528_2020_10025_MOESM1_ESM.pdf
Supplementary Information 1 Supplement Fig. S1 The nucleotide composition based on 96 Brachyura mitogenomes. Figure A is AT skewness graph, and figure B is GC skewness graph classified according to superfamilies. The abscissa axis is the total contents of AT or GC, and the vertical axis is AT skews or GC skews. Each colored dot represents a different superfamily (PDF 227 KB)
10528_2020_10025_MOESM2_ESM.pdf
Supplementary Information 2 Supplement Fig. S2 Secondary structures of 22 transfer RNA genes of M. abbreviatus. The dash (-) represents the Watson-crick pairing between two nucleotides (PDF 164 KB)
10528_2020_10025_MOESM3_ESM.pdf
Supplementary Information 3 Supplement Fig. S3 Hypothetical rearrangement mechanisms for Macrophthalmidae, Varunidae, Sesarmidae, and Dynomenidae by comparing with ancestor of Brachyura. Mitogenomes in red boxes represent genes which have changed. The red arrows represent tandem duplication and random loss and the deleted genes are marked in yellow boxes. The blue lines indicate the connection of the duplicate genes (PDF 191 KB)
10528_2020_10025_MOESM4_ESM.pdf
Supplementary Information 4 Supplement Fig. S4 The evolution of GOs in Majoidea and Potamoidea. The rearrangements in the GOs of Majoidea and Potamoidea species are investigated and depicted with respect to GOs of Brachyura. Mitogenomes in red and blue boxes represent genes which have changed. The red arrows represent tandem duplication random loss and transposition processes. The deleted genes are marked in yellow boxes (PDF 278 KB)
Rights and permissions
About this article
Cite this article
Xu, X., Wang, Q., Wu, Q. et al. The Entire Mitochondrial Genome of Macrophthalmus abbreviatus Reveals Insights into the Phylogeny and Gene Rearrangements of Brachyura. Biochem Genet 59, 617–636 (2021). https://doi.org/10.1007/s10528-020-10025-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-020-10025-8