Abstract
Many plant parasitic and entomopathogenic nematodes harbor specialized and obligate bacteria as well as viruses. Given their evolutionary persistence, such cryptic species are considered to play effective roles during their host/microbe interactions lifetime, including mutualistic, antagonistic, or yet unknown host effects. To exploit such associations in plant protection, a comprehensive view is needed linking basic evolutionary relationships to applied aspects. This requires identifying the benefit or impact that hosts, acting as pests or biocontrol agents, receive from their endosymbionts. Targeting endosymbionts that are vital for a beneficial nematode or a pest may open novel perspectives for the management of their performance and traits, such as virulence or response to plant defense reactions. Some hypotheses are proposed to develop advanced control strategies through emerging biotechnological approaches.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Key message
-
Endosymbionts represent a useful resource with high potential for sustainable nematode management
-
Some endosymbiotic bacteria sustain their host metabolism
-
Other endosymbionts, such as viruses and bacteria, negatively affect their hosts providing benefits to crops
-
Novel experimental efforts are needed for endosymbiont characterization
-
Technologies targeting or silencing endosymbionts may open new perspectives in sustainable plant protection
Background
A widespread and diverse range of microbial endosymbionts occurs in higher animals and plants in any habitat. Genomic and observational data consistently revealed the presence of a cryptic but complex microbiota also in many invertebrate lineages, including beneficial microorganisms, endosymbionts, and biocontrol agents that, in several cases, co-evolved with their hosts (Chaston et al. 2011; Shi et al. 2016; Dheilly et al. 2022). Given the time scale and dimensions of such evolutionary radiation, the diversity, role and impact of many invertebrate endosymbiotic species remain still partially unexplored.
Nematodes are among the most abundant organisms on the planet and play several, fundamental ecological roles. Plant parasitic nematodes (PPNs) represent a small fraction of the total nematode diversity. However, they cause severe damages accounting for up to 25% of global yield losses, with an annual economic impact estimated at ∼ 100 billion USD (Nicol et al. 2011). Most severe damages to agriculture are caused by sedentary root-knot and cyst nematodes, mostly found in crop** systems characterized by intensive monocultures. All PPNs are obligate pests, and their adaptations to parasitism gained benefits by acquiring genes from other species through processes such as horizontal gene transfer (Bird et al. 2015; Kikuchi et al. 2017). Similarly, the introgression of microorganisms with different endosymbiotic capabilities and metabolic behaviors represented an evolutionary turning point that characterized the radiations of many nematode lineages (Chaston et al. 2011; Shi et al. 2016).
A cryptic and diverse microbiota occurs in many nematode taxa. “Cryptic” herein refers to microbial species that are difficult to characterize, recognize or even identify. Nematodes play a direct role in the crop environment, acting as pests or insect pest regulators, such as PPNs or beneficial entomopathogenic nematodes (EPNs), respectively. The discovery of cryptic endosymbionts often resulted from direct microscopic observations, multigenic metabarcoding or NGS-based -omic studies. These observations highlighted the insurgence of specific traits of cooperation or conflicting interactions (Murfin et al. 2012; Baquiran et al. 2013; Martinson et al. 2020). PPNs and EPNs harbor a cohort of microorganisms (bacteria and/or viruses) underpinning complex relationships, influencing, in some cases, the evolution of their hosts and involved in various metabolic and/or pathogenic interactions (Shi et al. 2016; Ogier et al. 2023).
Despite efforts to identify the nematode microbiota and to understand such associations, much information is still lacking, given the phylum Nematoda complexity and spread. A deep insight into the evolution of these systems could be important also to control the most severe pests. Extensive studies on symbiosis, supported by complete genomic data, may reveal how such interactions and adaptations persisted and were influenced by multiple factors. For example, it is not yet known whether and how agricultural practices and plant physiology affect the relationships between endosymbionts and their host nematodes. Consequently, few information is available on the symbionts effects on the EPN efficacy and virulence or the PPN resistance to chemical treatments or to plant defensive metabolites.
Given their evolutionary persistence, such microbiota play highly effective roles for the hosts that were favored by selection, including mutualistic interactions. Endosymbiosis, however, involves the co-occurrence of antagonistic microorganisms, which are included in this review. Our goal was to update the most significant data available thus far on the topic for PPNs and EPNs, aiming at evaluating the impact of endosymbiotic microorganisms on plant production, and their potential in the regulation of pests. Basic issue is to identify most suitable symbiont/pest associations, harnessing such species to manage plant pests (either nematodes and insects).
Nematode endosymbiotic bacteria
Obligate vertically transmitted endosymbionts
Studies on nematode endobacteria mostly focused on their detection and genomic characterization. The discovery of nematode endosymbiotic bacteria often resulted from ultrastructural or taxonomic studies. Few data are available about their impact on the corresponding host biology and physiology or about their potential to mitigate the damage that their hosts induce on parasitized plants. In fact, the quantitative evaluation of the effects of such associations is difficult due to the paucity of suitable culturing methods for bacteria that remain in large part unclassified or at the “Candidatus” status. In recent years, however, multiple genomic approaches allowed more in-depth and in-host functional analyses. They were useful to identify and characterize such interactions, partially bypassing the need for cultivation (Brown et al. 2015, 2016).
Vertically transmitted endosymbionts are passed from a parental generation to the offspring, being associated with the juveniles during the egg or embryo formation. Among them are PPN endocellular symbionts of genera “Candidatus Cardinium” (Bacteroidetes) and Wolbachia (α-Proteobacteria) (Noel and Atibalentja 2006; Atibalentja and Noel 2008; Haegeman et al. 2009; Brown et al. 2016; Brown 2018; Haegeman et al. 2009; Tarlachkov et al. 2023). Further bacterial endosymbionts reported in PPNs are “Candidatus **phinematobacter” (Verrucomicrobia) and “Ca. **phinematincola” (Burkholderiales) found in **phinema spp. (Coomans et al. 2000; Vandekerckhove et al. 2000; Brown et al. 2015, 2016; Orlando et al. 2016; Palomares-Rius et al. 2021). These taxa likely represent a small fraction of the natural diversity of bacteria associated to nematodes, as other species and associations likely remain yet to be discovered and described.
Candidatus Cardinium
These Gram-negative, rod-shaped bacteria include intracellular endosymbionts present, with distinct lineages, among arthropods and nematodes. The bacteria grouped in “Ca. Cardinium” attracted interest as they can modify the host reproduction mode through induced parthenogenesis, feminization and cytoplasmic incompatibility (Penz et al. 2012; Tarlachkov et al. 2023). Bacterial cells similar to “Ca. Cardinium” were initially observed in the cyst nematodes Heterodera glycines and H. avenae (Endo 1979; Yang et al. 2017). Transmission electron microscopy images showed a similar endosymbiont in other cyst nematodes such as Globodera rostochiensis and H. goettingiana. The bacteria were localized in the ovary and oviduct wall cells and oocytes, suggesting a vertical transmission (Shepherd et al. 1973). The new taxon “Ca. Paenicardinium endonii” was then proposed for a similar endosymbiont found in second stage juveniles (J2) and adults of H. glycines (Noel and Atibalentja 2006). The bacterium was located in the J2 pseudocoelom, hypodermal chords and intestine, in oocytes, ovaries, spermatozoa, and intestine of both females and males, and the hypodermal chords of males. Electron microscopy images also confirmed the presence of microfilament-like structures in the bacterial cells with an unknown function (Endo 1979; Noel and Atibalentja 2006).
Tarlachkov et al. (2023) recently revised the genus “Ca. Cardinium” reporting the occurrence of these bacteria in twelve cyst nematode species from four genera of Heteroderidae, with a further new taxon in the root lesion nematode, Pratylenchus penetrans. Phylogenetic analyses confirmed five distinct evolutive branches associated with marine copepods, biting midges, mites, and cyst nematodes, adding two new groups associated with P. penetrans, and ostracods (Tarlachkov et al. 2023). These authors also sequenced a new “Ca. Paenicardinium” from H. humuli whose genome (1.05 Mb) appeared to be smaller than that of other nematode “Ca. Paenicardinium” genomes sequenced from H. glycines (NCBI acc. n. CP029619) and P. penetrans, although it exhibited a higher G + C content. Genome sequencing data from populations of Rotylenchus zhongshanensis showed, also in this species, a further taxon phylogenetically close to the “Ca. Paenicardinium” occurring in cyst and lesion nematodes (Guo et al. 2022).
Wolbachia
Bacteria of this genus belong to the α-Proteobacteria lineage. They are strongly associated with their hosts, including members from different evolutive branches. Species found in arthropods act as pathogens or affect the host reproductive biology. Wolbachia spp. play a fundamental nutritional role in their hosts. Nutritional mutualism between insects and Wolbachia strains has also been reported, evidencing the ability of the endosymbiont to synthesize vitamins that significantly contribute to the host fitness (Nikoh et al. 2014). In the bed bug Cimex lectularius, a gonad-associated Wolbachia (wCle) showed changes in the infection frequency and abundance that were linked to the host life stage. Such obligate mutualism involves the production by wCle of vitamins such as biotin and riboflavin, that are required by the host during its development (Moriyama et al. 2015; Fisher et al. 2018). Among Nematoda, Wolbachia species are found in filarial nematodes, in which they have a mutualistic role (Sironi et al. 1995; Manoj et al. 2021). Also, in Brugia malayi, the causal agent of filariasis in humans, the abundance of the bacterium was found to vary in relation to the host life stage (Fenn and Blaxter 2004). The discovery of Wolbachia endosymbiosys in filarial nematodes provided a first drug target to implement a successful human chemotherapy, based on various antibiotics (Hoerauf et al. 2000; Rao 2005; Bouchery et al. 2013). Such achievement is a practical example of the benefits derivable by the knowledge about endosymbionts, to be hopefully replicated with nematode pests.
The Wolbachia evolutionary history and biology have been extensively investigated (Werren 1997; Kaur et al. 2021) including the horizontal transmission events leading to insect host switch, not observed in filarial nematodes (Porter and Sullivan 2023). The presence of a Wolbachia-like endosymbionts in PPNs was observed while studying genes expressed by the burrowing nematode Radopholus similis, parasitizing banana plants. Studies of the R. similis genome highlighted approximately 1% of genes with a similarity to Wolbachia sequences (Haegeman et al. 2009; Jacob et al. 2008). The role of Wolbachia in R. similis is unknown. However, there is an indication that it provides the host with essential metabolites, required for the nematode survival. Recent phylogenetic analyses revealed a new Wolbachia also in P. penetrans populations that also hosted a Cardinium endosymbiont (Brown et al. 2016; Wasala et al. 2019; Kaur et al. 2021).
Verrucomicrobia
Molecular ecology studies on these bacteria highlighted their ubiquitous occurrence, with members often reported from rhizosphere soil analyzed in metabarcoding studies (Schlesner et al. 2006). Nematode-hosted Verrucomicrobia include “Ca. **phinematobacter”, a group of obligate intracellular endosymbionts of the dagger nematode **phinema spp. (Coomans et al 2000; Vandekerckhove et al. 2000). A strict, specific host association was reported for three species, namely “Ca. **phinematobacter brevicolli”, “Ca. **phinematobacter americanum” and “Ca. **phinematobacter rivesi”, found in X. brevicollum, X. americanum and X. rivesi, respectively (Vandekerckhove et al. 2000). The bacteria reside inside the epithelial cells of the nematode ovarian wall, and appear to influence their host reproductive mechanism. The 0.9 Mb genome of “Ca. **phinematobacter” has been fully sequenced (GenBank acc. n. CP012665), revealing genes with a fundamental mutualistic role in the biosynthesis of nematode-essential amino acids (aa) and nutrient hiring (Brown et al. 2015). Members of this clade appear particularly sensitive to their microhabitat. In juveniles, they are located in the gut epithelial cells, while in the eggs the bacteria are clustered at one of the poles. From this location they spread, during the embryo development, to all endodermal daughter cells, to guarantee the vertical transmission pathway (Coomans et al. 2000; Vandekerckhove et al. 2000; Brown et al. 2015). The host nematode genus includes economically important pests, some of which are vectors of plant viruses. However, there are no data available on the effect of Ca. **phinematobacter on the host vectoring capability or its plant parasitic feeding behavior.
Other bacterial endosymbionts
Recently, a new intracellular bacterium was observed in further **phinema species, as shown by a new 16S rRNA ribosomal gene sequence produced from X. pachtaicum, clustering in the family Burkholderiaceae (β-Proteobacteria). Observations showed that the bacteria were located inside the body of adult X. pachtaicum females, particularly in the ovaries and intestinal epithelium (Palomares-Rius et al. 2021). Although no direct observation was provided about its transmission route, the location in the ovaries likely suggests a vertical transmission mode.
Phoretic endosymbionts (horizontally transmitted)
Pasteuria
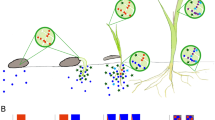
Some endosymbionts can be horizontally transmitted to other members of the same or different species through non-hereditary means. Horizontal transmission occurs from an infected host to a new uninfected one, irrespective of their generation time. Pasteuria spp. (Bacillales) are nematode parasites having a vegetative life cycle that occurs entirely within their hosts. As such, they may be considered endosymbiotic. Being effective biocontrol agents, they can regulate the density of many host nematode species in the rhizosphere environments. Pasteuria spp. include taxa parasitic in free-living nematodes or PPN. They are invariably characterized by a specific and narrow host range, and by the production of durable, cup-shaped endospores (Sayre et al. 1991; Sturhan et al. 2005; Stackebrandt 2014). The endospores are released in soil at the end of previous infection cycles and adhere to the nematode cuticle, acting as both infective and resting propagules (Fig. 1A). They show a long-term resistance to adverse conditions and allow the bacterium to persist in the soil environment outside the host body. The latter is the only trophic niche suitable for the Pasteuria vegetative growth. There are no available data about the factors that trigger endospore germination, which has been always observed after adhesion to the host (Fig. 1A).
Competitive exclusion of horizontally transmitted pathogenic endosymbionts parasitizing Meloidogyne incognita. Infective endospores of Pasteuria penetrans attached to the cuticle of a living and healthy juvenile stage (A) and on a nematode already parasitized by a yet unclassified pathogen (B, C), that leads to a complete host digestion (D). Scale bars: A, D = 10 μm; B = 5 μm; C = 1 μm
Although fastidious in nature, these bacteria have been produced in vitro and on larger scales on specific acidic media. Some isolates have been made available in commercial bioformulations tailored on targeted applications (Hewlett et al. 2004). The products include formulations based on endospores of Pasteuria penetrans for management of root-knot nematodes (Meloidogyne spp., RKN). Other species are P. nishizawae for biocontrol of the soybean cyst nematode (H. glycines, SCN), and P. usgae for the sting nematode (Belonolaimus longicaudatus). However, despite data produced from field trials in different agro-ecosystems (Luc et al. 2010; Crow et al. 2011), several aspects of the Pasteuria-host interactions still need to be investigated in detail to improve its efficacy in biocontrol, especially related to its very narrow host specificity.
Some Pasteuria spp. have been investigated for nematodes management through field and greenhouse assays (Verdejo-Lucas 1992; Atibalentja et al 1998; Gowen et al. 2008; Timper et al. 2016). Pausteria spp. host regulation occurs through a density-dependent relationship. The bacterium may persist in the rhizosphere environment for a long time, and was detected in the hosting nematode field populations up to two decades after its initial discovery (Ciancio et al. 2016).
Progress on knowledge about Pasteuria spp. biology has been achieved through the release of genome sequencing data. The partially annotated genome of P. penetrans (isolate RES148) from RKN, shows a reduced genome (2.64 Mb, GenBank acc. n. ERZ1024078) enriched in transposase and collagen genes, indicating a partial dependence on the host for basic metabolic pathways (Orr et al. 2018).
Genome sequencing efforts aimed at facilitating mass production by circumventing limitations related to the stringent metabolic requirements that make the Pasteuria industrial production challenging and limited to niche applications. In fact, apart from host specificity and biocontrol efficacy, few data have been produced on the physiology, biochemistry and metabolic requirements of Pasteuria spp. (Phani et al. 2018). Despite such limitations, these parasites have a huge potential in organic agriculture as well as in conventional nematode management, due to their obligate and host-specific parasitism. Assays with inundative treatments showed a remarkable reduction of target nematode densities, with an effect that was persistent in time (Cetintas and Dickson 2004).
The application potential of such nematode-associated bacteria appears far from being fully exploited, given the number of isolates and species (in the order of hundreds) resulting from their evolutive radiation and spatial spread. Further benefits include the high degree of host specificity and the null impact on other nematodes present in the rhizosphere (Gowen et al. 2008; Timper 2009), or the soil environment, together with the possibility of long-term storage at ambient temperature as durable and infective endospores.
Other bacteria
Other horizontally transmitted bacteria frequently were encountered in Italy in solanaceous crops. They include an unclassified and unculturable Gram-negative species pathogenic in J2 of M. incognita and other RKNs. Mature bacteria have a peculiar rod morphology resulting from four cells sequentially joined, yielding a kind of septa with empty and tapering apical cells (Fig. 1B–D). The bacterium proliferates after adhering germinating cells penetrate the J2, then completely digesting the host body content. Dead nematodes filled with cells release the bacteria after body rupture, which infects new J2 through passive adhesion in soil. Concomitant infections were observed in J2 already encumbered with P. penetrans endospores, leading to the competitive exclusion of the latter, as the bacterium life cycle appears faster and is already completed within the J2 in soil (Fig. 1B). The bacterium is fastidious, and no pure culture could be produced on most common nutritive media (Ciancio 2021).
Entomopathogenic nematodes and bacteria
Entomopathogenic bacteria (EPBs) are widely distributed and exhibit diverse host ranges, including EPNs (Steinernema, Heterorhabditis) and EPN-like nematodes (eg., Caenorhabditis, Oscheius). These associations vary from phoresis and commensalism to facultative or obligate parasitism (Tarasco et al. 2023). As a result of the mutualistic partnership between the nematode and its bacterium, pathogenesis is facilitated (Ogier et al. 2023).
Obligate EPBs, such as Photorhabdus and Xenorhabdus, complete their life cycles exclusively within the final insect host, while facultative ones (e.g., Serratia) can also grow outside the host. Endosymbionts Xenorhabdus and Photorhabdus establish a genus-specific association with the infective juveniles (IJs) of Steinernema and Heterorhabditis, respectively, obligate and lethal insect parasites. The genus Serratia is frequently associated with some nematode species of Oscheius, Caenorhabditis and other EPNs. They can be isolated, depending on the case, from the gut or cuticle of juveniles (Loulou et al. 2023; Oggier et al. 2023). Serratia comprises species and strains recognized for their entomopathogenicity, such as S. nematodiphila and S. marcescens. The latter species, together with Providencia rettgeri, are involved as bacterial mutualists in parasitism of the harlequin ladybird (Harmonia axyrid) by the parasitic nematode Parasitylenchus bifurcatus, inducing the expression of sex-specific host immune responses (Gegner et al. 2018).
Recent sequencing studies reported that the core symbiont is also associated, in EPNs and EPN-like nematodes, with a diverse bacterial community, referred to as the “second bacterial circle” (Ogier et al. 2023). The microbiota community includes species of several genera, such as Proteus, Ochrobactrum, Providentia, Acinetobacter, Pseudomonas, Alcaligenes, Stenotrophomonas, among others (see Ogier et al. 2020, 2023). This pathobiome plays a role in contributing to the nematode pathogenicity. However, the nematode-bacteria interaction is not always beneficial to the host. In some cases, the association has a detrimental effect on the nematode, as occurs in the phoretic and commensal relationship between Paenibacillus spp. and EPNs (Enright et al. 2003; El-Borai et al. 2005). The bacterium adheres to the IJ cuticle inhibiting its motility and, consequently, its efficiency to find the host (Enright and Griffin 2005).
EPBs Xenorhabdus and Photorhabdus stand out for their potential in pest management and other industrial applications (Abebew et al. 2022; Loulou et al. 2023). Currently, there are 31 and 22 valid species of Xenorhabdus and Photorhabdus, respectively (Machado et al. 2023a, b; Ritter et al. 2023). The free-living IJs of Steinernema and Heterorhabditis locate and penetrate the insect through natural openings or the cuticle. Subsequently, they migrate to the hemocoel and release their gut bacterial symbionts, which rapidly multiply and induce a septicemia, thus killing the host (Labaude and Griffin 2018). The bacteria possess the ability to produce numerous virulence factors, allowing them to evade the insect immune response and the host microbiota (Tobias et al. 2018). Additionally, they play a protective role by producing several antibiotic molecules that prevent insect invasion by other contaminating microorganisms (Bode 2009; Raja et al. 2021; Awori 2022). In this mutualistic association, the insect body becomes a suitable medium for the feeding and development of EPNs, which later feed on the bacteria and multiply in the insect cadaver, while the symbionts benefit from being transported by the IJs from one host to another.
Evolution and co-evolution
The nematode families Steinernematidae and Heterorhabditidae evolved at roughly the same time in the mid-Paleozoic, ca. 375 M years ago (Poinar 1983). However, although both families present similarities, their life cycle and association with EPBs are the result of convergent evolution (Chaston et al. 2011). They had separate origins (polyphyletic), Steinernema evolving from a “proto-Rhabditonema” ancestor in a terrestrial environment and Heterorhabditis from a “Pellioditis-like-ancestor”, in a coastal habitat (Poinar 1993).
Taxonomy and evolutionary history of EPN symbiotic bacteria have been re-assessed based on advances in molecular phylogeny (Sajnaga and Kazimierczak 2020). In contrast to their EPNs hosts, Xenorhabdus and Photorhabdus are phylogenetically more closely related to each other than to any other known species, being the genus Proteus the nearest neighbor. About 200–500 M years ago, a common ancestor of these bacteria was able to associate with Steinernema and Heterorhabditis (Sivaramakrishnan and Razia 2021). Subsequently, under selective pressures for maintaining this symbiosis, the ancestor evolved two distinct bacterial genera that established specific associations with their host (Chaston et al. 2011).
In most cases the symbiotic association is highly specific and provides an excellent model to understand EPN evolution (Sajnaga and Kazimierczak 2020). The host preference is highest in Steinernema, in which each species established a symbiosis with one species of Xenorhabdus, whereas Xenorhabdus spp. can be associated with different Steinernema species (Lee and Stock 2010; McMullen et al. 2017). Meanwhile, within the Heterorhabditis-Photorhabdus complex, multiple associations between EPNs and bacteria species can be observed (Maher et al. 2017; Abd-Elgawad 2021).
Co-phylogenetic analyses provide insights into the evolutionary dynamics and interactions linking nematodes and their symbiotic bacteria. Some multigene approaches revealed a notable congruence between the phylogenies of EPNs and EPBs, which suggests that co-speciation occurred in some lineages of the Steinernema/Xenorhabdus and Heterorhabditis/Photorhabdus complex (Maneesakorn et al. 2011; Bhat et al. 2019). However, other studies found no strict evidence of global co-evolution patterns (Lee and Stock 2010). Mismatches between the two phylogenetic trees can be attributed to factors such as host switching, in which EPBs switch between different nematode hosts, or incomplete host specificity, allowing EPBs to colonize additional nematode species. Host switching and incomplete specificity can enlarge the bacterium host range, thus yielding inconsistencies between the phylogenies of symbiotic partners (Maneesakorn et al. 2011).
The capacity for host switching enables bacterial symbionts to spread to different nematode taxa and adapt to new ecological niches, facilitating the dissemination of phylogenetically conservative traits (Sajnaga and Kazimierczak 2020). However, host switching can also have detrimental consequences on the nematode host, resulting in a decline in reproductive fitness, symbiont carriage, and virulence (McMullen et al. 2017). These authors stated that the fitness of both partners declines as the phylogenetic distance from the native association increases, suggesting that interaction specificity impacts the EPN virulence and longevity. In the symbiotic relationship, maintaining virulence is crucial for the reproductive fitness (Stock 2019), facilitating the successful transmission of both partners from one insect host to another. The phylogenetic reconstruction of different strains of Photorhabdus showed a general evolutionary trend towards an increase in virulence (Blackburn et al. 2016). Furthermore, studies have indicated that evolved, more virulent bacterial strains showed reduced production of bacteriocin (antimicrobial peptides that are effective vs other bacteria) and faster growth, compared to their ancestral populations, providing evidence that bacteriocin production carries significant costs at the population level (Bhattacharya et al. 2019).
Viruses
Plant pathogenic viruses vectored by nematodes
Nematodes viriome includes a number of plant pathogenic viruses that are vectored and transmitted to the host plants by a relatively low number of species. The link between plant viruses and nematodes is known since the first description of Grapevine Fanleaf Virus (GFLV) transmission by the longidorid nematode X. index (Hewitt et al. 1958). Following this discovery, most information and data have been produced on the transmission of Nepo- and Tobraviruses to plants by members of the nematode families Longidoridae and Trichodoridae (Schellenberger et al. 2011). This vectoring association is non-pathogenic for the nematodes. It has been the object of several reviews available in the current literature (Taylor and Robertson 1975; Singh et al. 2020).
Both Longidoridae and Trichodoridae are polyphagous ectoparasitic nematodes belonging to distinct evolutionary lineages (Dorylaimida and Triplonchida, respectively). Given their phylogenetic distance, the vector-virus association is considered to have appeared independently at least twice during the nematodes evolution. To date, twenty-four Longidoridae species are known to transmit twelve Nepovirus and one Sadwavirus, whereas all three Tobravirus are vectored by thirteen nematode species of genera Paratrichodorus and Trichodorus (Trichodoridae) (Decraemer and Robbins 2007; Bileva et al. 2009).
** the particles for years and even during long-term storage in soil (Tzortakakis 2023). GFLV and its vector are present in almost any grapevine producing region worldwide, and cause a severe crop decline with high economic losses. The GFLV particles acquired during feeding remain adsorbed on the inner lining of the stylet and oesophagus, where they are retained. The virus coat protein mediates the particles adsorption on the inner cuticle as well as their movement inside the plant infected tissues, thus being responsable of the vector specificity. The particles are transmitted when the nematodes feed on new roots but are lost at moult (Wang et al. 2002).
Other longidorid vectors of plant viruses are X. diversicaudatum, that transmits Strawberry latent ringspot sadwavirus (SLRV) and Arabis mosaic nepovirus (AMV), and X. rivesi, vector of Cherry raspberry leaf nepovirus (CRLV, Cheravirus), Tobacco ringspot nepovirus (TRSV) and Tomato ringspot nepovirus (TomSRV). **phinema rivesi showed a long-term retention of TomRSV particles for up to three years, with surviving individuals capable of transmitting the virus for two more years (Bitterlin and Gonsalves 1987). **phinema americanum sensu stricto is also vector of CRLV, TRV and TomSRV. Members of X. americanum sensu lato (non-European populations) transmit Peach rosette mosaic nepovirus (Bileva et al. 2009). Other longidorid vectors include Longidorus apulus and L. fasciatus, vectors of Artichoke italian latent nepovirus; L. elongatus, vector of Raspberry ringspot nepovirus (RRSV) and Tomato black ring nepovirus; L. arthensis, vector of Cherry rosette nepovirus and L. macrosoma, vector of RRSV (Brown et al. 1995). Paralongidorus maximus is vector of AMV, SLRV, CLRV and RRSV (Jones et al. 1981; Bileva et al. 2009).
Among Trichodoridae, species of Paratrichodorus and Trichodorus are vectors of Tobacco rattle tobravirus (TRV), Pea early-browning tobravirus and Pepper ringspot tobravirus (Harrison and Robinson 1986). Paratrichodorus pachydermus and Trichodorus primitivus are vectors of TRV, one of the causal agents of potato spraing disease (Brown et al. 1995; MacFarlane 2003).
Nematode viruses
Apart of plant viruses, molecular studies showed the occurrence of other virus lineages infecting nematode hosts, likely acting as pathogens or introgressed in the host genome as the result of ancestral insertions. Until recently the known interactions of viruses and nematodes were limited to a number of Caenorhabditis species (Félix et al. 2011; Franz et al. 2012, 2014; Frézal et al. 2019; Guo et al. 2014; Fujii and Wang 2023), an iridovirus found in the insect parasitic nematode Thaumamermis cosgrovei (Hess and Poinar 1985), an icosahedral virus particles found in Gastromermis sp. (Poinar and Hess-Poinar 1992), and to a virus associated with Capillaria hepatica, a nematode parasitic of mammals (Williams et al. 2019). A futher virus associated to the EPN S. ceratophorum was recently identified (Wang et al. 2022).
Virus detection during the routine examination of nematodes in light microscopy is not possible, as no specific symptom is associated to infections. Determining whether a nematode is infected by a virus is indeed a difficult task, requiring at least patient and consistent serial electron microscopy observations. Many PPNs, such as RKN or cyst nematodes, i.e., potato cyst nematodes (Globodera spp., PCN) or SCN, have a sedentary, parasitic life cycle and microscopic dimensions that make it difficult to observe stages in quantities sufficient to identify possible viral symptoms. Fortunately, new high-throughput sequencing methods (i.e., NGS) are facilitating the investigation of nematode-viral genomes and related discoveries (Kumar et al. 2017; Vieira et al. 2022). NGS methods produce massive amounts of data, require relatively low amounts of starting material, and allow many potentially infected individuals to be tested in a single assay (Posada-Cespedes et al. 2017; Pérez-Losada et al. 2020). Using this new approach, novel viral genomes have been discovered and identified from PPNs in the last years (Supplementary Table 1). DNA-based studies have in fact demonstrated the presence of viral lineages in the body of various PPN hosts, which likely act as pathogens or occur as the result of ancestral genome insertions.
There are few data on the effects of viruses on the parasitic fitness of nematodes, as well as on their tropism and on the mechanisms controlling transmission. The viriome of nematodes represents indeed a recent and new research field. Due to the ecological complexity of the rhizosphere food webs and, in a broader sense, of many agricultural systems, the study of viral infections in PPN is challenging. The occurrence of PPN infecting viruses suggests a potential for biotechnological applications to manage most severe pests in a sustainable way. For example, field and greenhouse data indicated that SbCNV-5 may affect SCN growth, acting as a biological control agent (Ruark et al. 2017). Visible, debilitating symptoms were reported for RKN infected by a virus-like pathogen, although no viral particles were visualized (Loewenberg et al. 1959). A reduced fitness was reported in specimens of the free-living C. elegans infected by the Orsay virus, with changes in sexual behavior not observed in healthy nematodes (Frézal et al. 2019; van Sluijs et al. 2021).
Soybean Cyst Nematode (SCN) viruses
Heterodera glycines is a severe parasite of soybean with a complex interaction with its host (Bandara et al. 2020). Bioinformatic investigations and SCN transcriptome assembly revealed nearly complete genomes of both negative- and positive-sense, single-stranded RNA (ssRNA) viruses. SCN viruses are not integrated into the host genome. Detection of positive-strand SCN viruses indicated that they could replicate in J2. Such viruses were found in a greenhouse population of a single-cyst descent SCN line (Colgrove and Niblack 2008) and in field SCN populations (Bekal et al. 2014). SCN virus infections are persistent. RNA samples extracted from SCN over a 4-years period indicated that the viral infections were long-lived and stable in the populations examined. Heterodera glycines-associated viruses have been also reported within populations of the clover cyst nematode H. trifolii and in the sugar beet cyst nematode H. schachtii (Ruark et al. 2017).
ScNV
ScNV contains five open reading frames (ORFs) coding for a nucleoprotein (N), a phosphoprotein (P), a matrix protein (M), a glycoprotein (G) and a RNA-dependent RNA polymerase (RdRp) large, non structural protein (L). ScNV genome is similar to the Midway virus (MIDWV) (Takahashi et al. 1982). MIDWV and Nyamanini virus (NYMV) are closely related members of genus Nyavirus, in a distinct taxonomic unit in the family Bornaviridae, but in a distinct taxonomic unit (Mihindukulasuriya et al. 2009). Neighbour-joining phylogenetic analysis of the RdRp aa sequences showed that ScNV is similar to viruses in the order Mononegavirales, which are enveloped ssRNA viruses, with monopartite genomes of negative polarity.
ScRV
The ScRV genome contains five predicted ORFs similar to ScNV in size (Bekal et al. 2011). The predicted aa sequence of the largest ORF (V) in the ScRV genome is 46% similar to the RdRp or the Northern cereal mosaic virus (NCMV) L protein. Phylogenetic analysis of RdRp sequence showed the closest relationship of ScRV with the order Mononegavirales, member of the family Rhabdoviridae, genus Cytorhabdovirus.
ScPV
The ScPV genome, contains a single large ORF, that is closely related to enveloped viruses in the family Bunyaviridae, genus Phlebovirus (Bekal et al. 2011). Phleboviruses have tripartite negative-sense ssRNA genomes. However, due to the less conserved products of the shorter M and S RNAs, only the L segment of the ScPV genome, containing the RdRp coding sequence, could be detected. ScPV groups with the type virus of this family (Bunyamwera virus) and is very similar to the Uukuniemi virus (UUKV) (Flick et al. 2002).
ScTV
The genome of ScTV includes in a single ORF. It is related to Rice stripe virus (RSV) and Rice grassy stunt virus (RGSV), both members of genus Tenuivirus. Like tenuiviruses, the ScTV L-protein is larger than the L proteins of most phleboviruses (Toriyama et al. 1994).
SbCNV-5c
The genome of the Soybean Cyst Nematode virus-5 (SbCNV-5c) encodes a single, 6004 aa polyprotein (Bekal et al. 2014). Phylogenetic analysis performed on conserved regions of the RdRp and RNA helicase indicated that SbCNV-5 is close to viruses in the genus Pestivirus. SbCNV-5 has been detected in greenhouse and field populations of SCN and in all four host stages: eggs, J2s, adult males and females (Bekal et al. 2014; Ruark et al. 2017). SbCNV-5 is not integrated into the SCN genome (Bekal et al. 2014).
SCN NLV
This virus belongs to order Mononegavirales (family Nyamiviridae). SCN NLV encodes a predicted nucleoprotein N, a phosphoprotein P, a glycoprotein G, the RdRp and two furher ORF-encoded proteins, with no attributable functionality due to the absence of sequence similarity (Ruark et al. 2018). SCN NLV is close to SbCNV-1 but it is not classified in the genus Socyvirus. The SbCNV-1 and SCN NLV genomes differ by approx. 50%, indicating that SCN NLV likely represents a new genus or that the species boundaries in this lineage should be expanded. SCN NLV has been sequenced from ten SCN populations and from H. trifolii (Ruark et al. 2018).
SCN BLV
SCN BLV is a futher RNA virus from order Bunyavirales, characterized by a negative-sense multipartite genome. Just like SCN NLV, SCN BLV also infects other arthropods, such as crustaceans and insects (Shi et al. 2016). Its polymerase sequence clusters with four other nematode host viruses. Detection with qRT-PCR and sequencing of its RdRps confirm the virus occurrence in nature as well as in the clover cyst nematode (Ruark et al. 2018).
Potato Cyst Nematode (PCN) viruses
The PcRV was discovered in populations of the PCN G. pallida through high-throughput sequencing. Its reverse complement strand includes five ORFs, with proteins varying from 180 (ORF 3) to 2180 (ORF 5) aa. This virus shows untranslated regions (UTR) of 156 and 641-nt at the 5’ and 3’ termini of the positive strand, respectively. ORFs include putative genes for a nucleoprotein N, a phosphoprotein P, an unknown gene, a glycoprotein G and a putative RdRp or large non-structural protein L (Kud et al. 2022). The largest ORF 5-encoded protein showed similarities with the L-proteins (RdRps) of rhabdoviruses. Phylogenetic analyses based on the L protein sequences placed PcRV close to ScRV in a distinct lineage, within an unclassified rhabdoviruses subfamily. Kud et al. (2022) proposed a new genus including both viruses from PCN and SCN, classifying PcRV and ScRV in the subfamily Gammanemrhavirus. PcRV was detected in eggs, J2, and females, likely indicating a vertical transmission. Infection appeared persistent and stable in the nematode populations examined, lasting at least 5-years.
Transcriptome data of G. pallida and G. rostochiensis revealed a positive-sense viral RNA genome named PCN PLV. The virus belongs to Picornavirales and produces a unique, predicted polyprotein (Ruark et al. 2018). PCN PLV does not show similarity with other viruses and requires an appropriate classification into a new genus. Proteases, helicases, and RdRps show conserved motifs of picorna-like viruses.
Sugar beet cyst nematode (SBCN) virus
Single-stranded positive-sense RNA viruses named SBCNV1 and SBCNV2, were predicted from analysis of the H. schachtii transcriptome (Lin et al. 2018). The SBCNV2 genome is organized in S, M and L segments, similar to the genome segments of members of some Bunyavirales.
SBCNV1 encodes a domain-containing polyprotein of members of Picornavirales. SBCNV1 was present in both eggs and J2 of SBCN, possibly indicating a vertical transmission. A negative-strand of SBCNV1 RNA was detected, indicating that replication of this virus occurs in H. schachtii (Lin et al. 2018). The SBCNV1 polyprotein contains domains of two rhinovirus (Rhv)-like picornavirus capsid proteins, a cricket paralysis virus (CRPV)-like (VP2-like) capsid, a RNA helicase, a peptidase-C3 and a RdRp, organized similarly to the genomes of Picornavirales (Le Gall et al. 2008). Phylogenetic analysis based on the RdRp conserved aa sequence domains placed SBCNV1 together with PCNPV near to members of families Iflaviridae and Secoviridae. The very low aa identities of SBCNV1 and PCNPV with other Picornavirales members raised the possibility of a new taxonomical classification within the Picornavirales (Lin et al. 2018).
Root lesion nematode (RLN) virus
The genome of RLNV1, a positive-sense single-stranded RNA, was discovered in two independent pools of transcriptomic datasets from the root lesion nematode P. penetrans (Thies et al. 1995). The presence of the virus has been confirmed in adults of both sexes but not in J2 or eggs. An hybridization assay, using an antisense probe of RLNV1m, detected the virus in the infected nematodes near to the esophageal glands (Vieira and Nemchinov 2019). Viral replication in RLN was also confirmed by PCR assay using a negative-strand-specific. RLNV1 belongs to the order Picornavirales. It consisted of a mono-cistronic genome with an ORF encoding a single, large polyprotein (Vieira and Nemchinov 2019). Phylogenetic analyses based on conserved RdRp domains placed RLNV1 in a distinct clade close to PCN PLV and SBCNV1, likely indicating the occurrence of a new family within the order Picornavirales. Furthermore, the low-coverage alignments (around 5%) with nucleotide sequences of SBCNV1 and PCNPLV suggest that RLNV1 likely represents a new genus (Koonin et al. 2008). The virus showed a widespread prevalence in North America and a low genetic variability (Vieira et al. 2020).
Pine wood nematode (PWN) endogenous viral element
eBxnv-1
The genome of the PWN, Bursaphelenchus xylophilus, a severe forest pest, includes an endogenous nodavirus-related sequence (eBxnv-1) (Cotton et al. 2016). This sequence is embedded in a degraded long terminal repeats (LTR) retrotransposon, suggesting that the nematode genome integrated this positive-sense viral RNA in a single exon gene, encoding a 570 aa hypothetical protein. This is similar to the RdRp (PS5057) of positive ssRNA viruses, and to the DNA/RNA polymerases superfamily (HMM SSF56672) (Cotton et al. 2016). NCBI Blast analysis for this protein in Genbank showed similarity only to RdRps of nodaviruses. Phylogenetic data showed its relationship to RDRp of nodaviruses and close links to a subgroup of arthropod-infecting alphanodaviruses found in Lepidoptera. However, electron microscopy observations were unable to detect any virus particle in a nematode tissue homogenate. Expression analysis of eBxnv-1 and LTR element (including the nodavirus-derived ORF), showed that they are expressed, although at a low level, during the whole B. xylophilus life-cycle, with no significant difference among developmental stages. Cotton et al. (2016) proposed the introgression of eBxnv-1 in B. xylophilus as an event where a RNA1 transcript, encoding the RdRp of a nematode nodavirus, infected the B. xylophilus germ line. Its reverse transcription in the cellular environment was likely followed by an LTR retroelement non-homologous recombination, leading to its insertion in the host genome. The B. xylophilus BUX.s01281.240, eBxnv-1 and BUX.s01281.242 genes are deposited in NCBI GenBank with acc. n. LC158686.
Entomopathogenic nematode virus
A dsRNA virus, named Steinernema ceratophorum partitivirus 1 (ScPV-1), was isolated from the entomopathogenic nematode, S. ceratophorum (Wang et al 2022). The positive-sense genome of ScPV-1 is composed of two segments of dsRNA1 and dsRNA2 with poly(A) tails at their 3’ termini (Supplementary Table 1). ScPV-1 belongs to the family Partitiviridae. Phylogenetic analysis of the putative RdRp confirmed that ScPV-1 is a new member of genus Betapartitivirus.
Future perspectives and novel research paths
Reports and related data on the biology of several endosymbionts are scattered in the scientific literature and gene databases, often resulting as indirect fallouts of research projects having a different goal. A more comprehensive view is needed, in relation to the potential of endosymbiotic associations and their links with crop productivity. Linking data derived from a basic evolutionary perspective to more applied aspects of plant protection requires the identification of the benefit that pests gain by their endosymbionts. A second aspect concerns the identification of host adaptation/virulence mechanism deployed by pathogenic endosymbionts to evaluate their effective value as targets or tools for PPNs or insect pests management.
The quantitative assessment of the impact of endosymbionts on crop productivity and pest regulation is of interest to achieve more sustainable plant protection methods and pest management approaches. The nutritional benefits that endosymbiotic associations provide to their hosts, for example, are only partially known and mostly for EPNs (Brown et al. 2015; Raja et al. 2021). There is in fact still a knowledge gap for several PPNs-endosymbiont associations. Having passed the filter of evolution and selection, many endosymbionts, individually or in consortia, provide consistent and fundamental crop ecosystem services directly involved both in pest fitness or regulation. Many of them i.e., Pasteuria spp., are known as effective PPN biological control agents in natural conditions, ensuring a sustainable stability of yields. However, the performance of their in vitro or industrial production still represents a factor limiting their widespread exploitation and application.
Initially, the interest in EPBs primarily focused on their symbiotic associations with EPNs. However, the current emphasis has shifted to their symbionts. The bioactive metabolites produced by these symbionts have a broad spectrum of applications, capable of killing harmful insects and PPNs, or microorganisms such as protozoa, bacteria and fungi (Abd-Elgawad 2022; Gulsen et al. 2022). EPBs secrete diverse arrays of bioactive compounds, including antibiotics, enzymes, bacteriocins, and toxins (see eg., Abd-Elgawad 2022; Parihar et al. 2022; Kallali et al. 2024). These secondary metabolites can also serve as valuable sources for new pesticide or drug compounds (Parihar et al. 2022). They may act as lead molecules in the development of alternative solutions to replace existing ones, with applications in agriculture, pharmaceuticals, and industry (Cimen et al. 2022; Vicente-Díez et al. 2023a).
The use of EPBs as biocontrol agents is an attractive and promising approach in sustainable agricultural systems to manage a wide range of plant pests and pathogens. EPBs can serve as effective tools for crop protection, acting as standalone pesticides with versatile formulations of either the bacteria themselves or their bioactive metabolites, and can be applied in various forms such as pellets, powder, spray, suspension, or supernatant (Abd-Elgawad 2021, 2022). They have shown efficacy against different plant pests, including PPNs (see eg., Kepenekci et al. 2016; Caccia et al. 2018; Sayedain et al. 2019; Kusakabe et al. 2022), insects (see eg., Shawer et al. 2018; Adithya et al. 2020; Yüksel et al. 2023; Vicente-Díez et al. 2023b, c), and fungi (see eg., Chacón-Orozco et al. 2020; Cimen et al. 2021; Gulcu 2022). The results of EPB applications are promising for simultaneously controlling different plant pathogens and insect pests. In addition to this, there is the possibility of applying consortia of EPBs, even with other biocontrol agents, achieving efficient results against insect pests (Spescha et al. 2023). However, additional research is imperative to optimize their utilization, encompassing a comprehensive understanding of their mode of action. Other goals include the development of effective formulations, and the identification of optimal application methods for field use. To make substantial progress in crop protection, these bacteria should be integrated into holistic crop management strategies (Abd-Elgawad 2022).
A possibility for PPN management relies in their gene modification. This approach is, however, still at an early stage due to various obstacles (Kranse et al. 2021). However, viruses can be considered as possible tools for such modifications, as already shown for animal parasitic nematodes (Hagen et al. 2021). SbCNV-5 appears as a candidate viral vector or for use, as infectious RNA, to manipulate expression of H. glycines genes. The relatively small, positive-sense RNA genome of SBCNV1, compared to other RNA viruses that infect PPNs, also makes it an ideal candidate for future molecular studies. These could potentially lead to new measures for management of SBCN and/or of other related pests. Future research on the impact of nematode viruses on host multiplication and virulence will help explore their potential as a new strategy for management and control of most PPNs.
Understanding the ecological aspects of the triple interaction between bacteria-EPN-insect host is also crucial for biocontrol and pest regulation. For example, insects have developed defensive strategies to resist the attack of their natural enemies, including the sequestration of secondary metabolites from the plants on which they feed (Erb and Robert 2016; Beran and Petschenka 2022). This protective effect is achieved by selective stabilization and reactivation of toxins targeting different stages of the infection process of EPNs and their symbiont bacteria, suppressing parasitism (Robert et al. 2017). For that reason, to enhance the efficacy of the EPN-bacteria complex in integrated pest management, different breeding techniques are being explored, including bacterial and EPN strain engineering (Machado et al. 2020; Abd-Elgawad 2023).
The integration in plant protection of newly emerging biotechnologies such as the use of microRNAs interference or micropeptides may yield benefits in the development of novel, low impact management strategies (Lauressergues et al. 2015). Silencing one of more genes active in the endosymbiotic associations that sustain a PPN survival may represent a future research objective. Efficient metabolism and gene expression are in fact required by endosymbionts to provide a nutritional benefit for their hosts, as shown by i.e., PPNs-associated Verrucomicrobia. However, most literature data deal with insect and bacteria associations, and a few afford the study of PPNs biology in terms of endosymbionts efficiency and host metabolism integration.
Methods in plant management through microRNAs and micropeptides have recently emerged as a fertile research field, with a potential in sustainable pest control (Lauressergues et al. 2015; Badola et al. 2022; Erokhina et al. 2023). The application of gene interference in endosymbiotic associations represents, hence, an attractive endeavor, given the severe impact that some species (i.e., RKN, SCN) have on yields, worldwide. Experimental data are, however, required to understand how to (i) determine the key metabolic pathways deployed by endosymbionts and (ii) interfere with their biology to build alternative and sustainable crop protection strategies.
Genetic engineering and manipulation technologies were applied to both Xenorhabdus and Photorhabdus. The insertion of a tetracycline-inducible promoter activated a gene cluster with a non-ribosomal peptide synthase, allowed the identification of novel secondary metabolites (Yin et al. 2015). These approaches are promising, as they can identify new metabolites such as antimicrobial peptides or other toxins, that may have a potential and impact in pest management.
A promising research field also concerns the interactions among different endosymbionts present in the same host, including the mechanisms of competitive exclusion, or the molecular mechanisms deployed by vertically transmitted bacteria in the interaction with their hosts (Vandekerckhove et al. 2000; Palomares-Rius et al. 2021). In particular, it is still unclear how endosymbiotic bacteria may evade the nematode defense system and related enzymes, i.e. lysozyme. EPBs in fact do not induce harm to, and are tolerated by, their EPNs phoretic hosts (Boehnisch et al. 2011). Host defensive mechanisms mediated by symbionts include the improvement of the host metabolic capabilities and vigor vs pathogens and parasites, together with chemical defense, competitive exclusion or immune system stimulation (Flórez et al. 2015). Defensive mechanisms are active also in nematodes. However, there are still a few studies focusing on evolutionary conserved mechanisms such as the innate immunity (Kurz and Ewbank 2000; Wang et al. 2019) or the occurrence and role of antimicrobial peptides. These are widespread among invertebrates in which they play an important defensive role. The antimicrobial peptides identified in nematodes include the neuropeptide-like proteins found in C. elegans (McVeigh et al. 2008), and cecropins, nemapores, and different lysozymes found in animal or plant parasitic species (Fanelli et al. 2008; Tarr 2012; Wang et al. 2019).
Lysozyme-mediated protection from invading bacteria appears particularly relevant. In rhabditids, lysozyme genes are expressed in the intestine, where their products act against pathogenic bacteria. However, the intestine is also the transit and storage environment of EPBs (Boehnisch et al. 2011). Different hypotheses may be formulated about the resistance or avoidance deployed by EPBs vs the lysozyme molecules (if any) produced by hosting EPNs. These include the possibility of a silencing effect exerted by EPBs on the expression of the host lysozyme genes, or the functional loss of lysozyme-encoding genes. In both cases a hypothetical protective role of EPBs vs other nematode pathogenic bacteria may be postulated, to compensate for such eventual host impairment. EPBs are known to produce a broad arsenal of antibiotics and bioactive compounds, acting as a chemical defense from bacteria, fungi or other invertebrates such as ants, that invade the insect cadaver in search of a food source (Flórez et al. 2015). However, no data are available about the production and release of such effectors also within the EPNs digestive tract. Lysozyme coding genes are present in other rhabditids. A BLASTx search in NCBI database using C. elegans lys-1 (locus CELE.Y22F5A.4.1) and lys-2 (CELE.Y22F5A.5.1) showed highest identities with genes in Caenorhabditis spp. and other rhabditids (Diploscapter pachys, Helicephalobus sp. and Pristionchus pacificus), or Strongylida. However, no significantly similar sequences could be retrived from Steinernema nor Heterorhabditis when blasting lys-1 and lys-2 sequences vs their genomes, apart of some fragments (data not shown). This incongruence deserves further attention, as it suggests a possible loss or degeneration of lysozyme encoding genes in EPNs.
In conclusion, the study of the nematode cryptic microbiome may yield new hypotheses and perspectives for future research work. These include the quantitative analysis of the endosymbionts impact on crops, encompassing not only plant parasitic nematodes but also the potential utilization of bacteria for controlling plant pests. A further aspect is the definition of gene silencing approaches to harm pests or improve the efficacy of biocontrol agents. Finally, a detailed insight on the symbiont/host/plant interactions may yield useful knowledge on fundamental pest traits such as hyper- or hypo-virulence, including the defense gene pathways activated by plant or insect hosts.
Availability of data and materials
The datasets used and/or sequence data analyzed during the current review are available in NCBI, from where genome data may be retrieved, or in the cited articles.
References
Abd-Elgawad MMM (2021) Photorhabdus spp.: an overview of the beneficial aspects of mutualistic bacteria of insecticidal nematodes. Plants 10:1660. https://doi.org/10.3390/plants10081660
Abd-Elgawad MMM (2022) Xenorhabdus spp.: an overview of the useful facets of mutualistic bacteria of entomopathogenic nematodes. Life 12:1360. https://doi.org/10.3390/life12091360
Abd-Elgawad MMM (2023) Optimizing entomopathogenic nematode genetics and applications for the integrated management of horticultural pests. Horticulturae 9:865. https://doi.org/10.3390/horticulturae9080865
Abebew D, Sayedain FS, Bode E, Bode HB (2022) Uncovering nematicidal natural products from Xenorhabdus bacteria. J Agric Food Chem 70:498–506. https://doi.org/10.1021/acs.jafc.1c05454
Adithya S, Shivaprakash MK, Sowmya E (2020) Evaluation of insecticidal activity of entomopathogenic bacteria Photorhabdus and Xenorhabdus against shoot and fruit borer Earias vittella (Lepidoptera: Noctuidae) of vegetable crops. J Entomol Zool Stud 8:2343–2348. https://doi.org/10.22271/j.ento.2020.v8.i4aj.7468
Atibalentja N, Noel G (2008) Bacterial endosymbionts of plant-parasitic nematodes. Symbiosys 46:87–93
Atibalentja N, Noel GR, Liao TF, Gertner GZ (1998) Population changes in Heterodera glycines and its bacterial parasite Pasteuria sp. in naturally infested soil. J Nematol 30:81–92
Awori RM (2022) Nematophilic bacteria associated with entomopathogenic nematodes and drug development of their biomolecules. Front Microbiol 13:993688. https://doi.org/10.3389/fmicb.2022.993688
Badola PK, Sharma A, Gautam H, Trivedi PK (2022) MicroRNA858a, its encoded peptide, and phytosulfokine regulate Arabidopsis growth and development. Plant Physiol 189:1397–1415. https://doi.org/10.1093/plphys/kiac138
Bandara AY, Weerasooriya DK, Bradley CA, Allen TW, Esker PD (2020) Dissecting the economic impact of soybean diseases in the United States over two decades. PLoS ONE 15:e0231141. https://doi.org/10.1371/journal.pone.0231141
Baquiran JP, Thater B, Sedky S, De Ley P, Crowley D, Orwin PM (2013) Culture-independent investigation of the microbiome associated with the nematode Acrobeloides maximus. PLoS ONE 8:e67425. https://doi.org/10.1371/journal.pone.0067425
Bekal S, Domier LL, Gonfa B, McCoppin NK, Lambert KN, Bhalerao K (2014) A novel flavivirus in the soybean cyst nematode. J Gen Virol 95:1272–1280. https://doi.org/10.1099/vir.0.060889-0
Bekal S, Domier LL, Niblack TL, Lambert KN (2011) Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J Gen Virol 92:1870–1879. https://doi.org/10.1099/vir.0.030585-0
Beran F, Petschenka G (2022) Sequestration of plant defense compounds by insects: from mechanisms to insect–plant coevolution. Ann Rev Entomol 67:163–180. https://doi.org/10.1146/annurev-ento-062821-062319
Bhat A, Chaubey A, Půža V (2019) The first report of Xenorhabdus indica from Steinernema pakistanense: co-phylogenetic study suggests co-speciation between X. indica and its steinernematid nematodes. J Helminthol 93(1):81–90. https://doi.org/10.1017/S0022149X17001171
Bhattacharya A, Toro Díaz VC, Morran LT, Bashey F (2019) Evolution of increased virulence is associated with decreased spite in the insect-pathogenic bacterium Xenorhabdus nematophila. Biol Lett 15:20190432. https://doi.org/10.1098/rsbl.2019.0432
Bird DM, Jones JT, Opperman CH, Kikuchi T, Danchin EG (2015) Signatures of adaptation to plant parasitism in nematode genomes. Parasitol 142(Suppl 1):S71-84. https://doi.org/10.1017/S0031182013002163
Bileva T, Choleva B, Hockland S, Ciancio A (2009) Management of virus-transmitting nematodes with special emphasis on South-east Europe. In: Ciancio A, Mukerji KG (eds) Integrated management of fruit crops and forest nematodes. Springer, Dordrecth, pp 215–252. https://doi.org/10.1007/978-1-4020-9858-1_9
Bitterlin MW, Gonsalves D (1987) Spatial distribution of **phinema rivesi and persistence of tomato ringspot virus and its vector in soil. Plant Dis 71:408–411. https://doi.org/10.1094/PD-71-0408
Blackburn D, Wood PL, Burk TJ, Crawford B, Wright SM, Adams BJ (2016) Evolution of virulence in Photorhabdus spp., entomopathogenic nematode symbionts. Syst Appl Microbiol 39:173–179. https://doi.org/10.1016/j.syapm.2016.02.003
Bode HB (2009) Entomopathogenic bacteria as a source of secondary metabolites. Curr Opin Chem Biol 13:224–230. https://doi.org/10.1016/j.cbpa.2009.02.037
Boehnisch C, Wong D, Habig M, Isermann K, Michiels NK, Roeder T, May RC, Schulenburg H (2011) Protist-type lysozymes of the nematode Caenorhabditis elegans contribute to resistance against pathogenic Bacillus thuringiensis. PLoS ONE 6:e24619. https://doi.org/10.1371/journal.pone.0024619
Bouchery T, Lefoulon E, Karadjian G, Nieguitsila A, Martin C (2013) The symbiotic role of Wolbachia in Onchocercidae and its impact on filariasis. Clin Microbiol Infect 19:131–140
Brown AMV (2018) Endosymbionts of plant-parasitic nematodes. Ann Rev Phytopathol 56:225–242. https://doi.org/10.1146/annurev-phyto-080417-045824
Brown AMV, Howe DK, Wasala SK, Peetz AB, Zasada IA, Denver DR (2015) Comparative genomics of a plant-parasitic nematode endosymbiont suggest a role in nutritional symbiosis. Genome Biol Evol 7:2727–2746. https://doi.org/10.1093/gbe/evv176
Brown AMV, Wasala SK, Howe DK, Peetz AB, Zasada IA, Denver DR (2016) Genomic evidence for plant-parasitic nematodes as the earliest Wolbachia hosts. Sci Rep 6:34955. https://doi.org/10.1038/srep34955
Brown DJF, Robertson WM, Trudgill DL (1995) Transmission of viruses by plant nematodes. Annu Rev Phytopathol 33:223–249. https://doi.org/10.1146/annurev.py.33.090195.001255
Caccia M, Marro N, Rondan Dueñas J, Doucet ME, Lax P (2018) Effect of the entomopathogenic nematode-bacterial symbiont complex on Meloidogyne hapla and Nacobbus aberrans in short-term greenhouse trials. Crop Prot 114:162–166. https://doi.org/10.1016/j.cropro.2018.07.016
Cetintas R, Dickson DW (2004) Persistence and suppressiveness of Pasteuria penetrans to Meloidogyne arenaria race 1. J Nematol 36:540–549
Chacón-Orozco JG, Bueno CJ, Shapiro-Ilan DI, Hazir S, Leite LG, Harakava R (2020) Antifungal activity of Xenorhabdus spp. and Photorhabdus spp. against the soybean pathogenic Sclerotinia sclerotiorum. Sci Rep 10(1):20649. https://doi.org/10.1038/s41598-020-77472-6
Chaston JM, Suen G, Tucker SL, Andersen AW, Bhasin A, Bode E, Bode HB, Brachmann AO, Cowles CE et al (2011) The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS ONE 6(11):e27909. https://doi.org/10.1371/journal.pone.0027909
Ciancio A (2021) Observations on a novel bacterial pathogen of root-knot nematodes (Meloidogyne spp.). Pathogens 10:1226. https://doi.org/10.3390/pathogens10101226
Ciancio A, Roccuzzo G, Ornat C (2016) Regulation of the citrus nematode Tylenchulus semipenetrans by a Pasteuria sp. endoparasite in a naturally infested soil. BioContr 61:337–347. https://doi.org/10.1007/s10526-015-9704-1
Cimen H, Touray M, Gulsen SH, Erincik O, Wenski SL, Bode HB, Shapiro-Ilan D, Hazir S (2021) Antifungal activity of different Xenorhabdus and Photorhabdus species against various fungal phytopathogens and identification of the antifungal compounds from X. szentirmaii. Appl Microbiol Biotechnol 105:5517–5528. https://doi.org/10.1007/s00253-021-11435-3
Cimen H, Touray M, Gulsen SH, Hazir S (2022) Natural products from Photorhabdus and Xenorhabdus: Mechanisms and impacts. Appl Microbiol Biotechnol 106:4387–4399. https://doi.org/10.1007/s00253-022-12023-9
Colgrove AL, Niblack TL (2008) Correlation of female indices from virulence assays on inbred lines and field populations of Heterodera glycines. J Nematol 40:39–45
Coomans A, Vandekerckhove TTM, Claeys M (2000) Transovarial transmission of symbionts in **phinema brevicollum (Nematoda: Longidoridae). Nematol 2:443–449
Cotton J, Steinbis S, Yokoi T, Tsai IJ, Kikuchi T (2016) An expressed, endogenous Nodavirus-like element captured by a retrotransposon in the genome of the plant parasitic nematode Bursaphelenchus xylophilus. Sci Rep 6:39749. https://doi.org/10.1038/srep39749
Crow WT, Luc JE, Giblin-Davis RM (2011) Evaluation of Econem, a formulated Pasteuria sp. bionematicide, for management of Belonolaimus longicaudatus on golf course turf. J Nematol 43:101–109
Decraemer W, Robbins RT (2007) The who, what and where of Longidoridae and Trichodoridae. J Nematol 39:295–297
Dheilly NM, Lucas P, Blanchard Y, Rosario K (2022) A world of viruses nested within parasites: unraveling viral diversity within parasitic flatworms (Platyhelminthes). Microbiol Spect 10(10):1128. https://doi.org/10.1128/spectrum.00138-22
El-Borai FE, Duncan LW, Preston JF (2005) Bionomics of a phoretic association between a putative Paenibacillus sp. and entomopathogenic nematode Steinernema diaprepesi. J Nematol 37:18–25
Enright MR, McInerney JO, Griffin CT (2003) Characterization of endospore-forming bacteria associated with entomopathogenic nematodes, Heterorhabditis spp., and description of Paenibacillus nematophilus sp. nov. Int J Syst Evol Microbiol 53:435–441. https://doi.org/10.1099/ijs.0.02344-0
Enright MR, Griffin CT (2005) Effects of Paenibacillus nematophilus on the entomopathogenic nematode Heterorhabditis megidis. J Invertebr Pathol 88:40–48. https://doi.org/10.1016/j.jip.2004.10.002
Endo BY (1979) The ultrastructure and distribution of an intracellular bacterium-like microorganism in tissue of larvae of the soybean cyst nematode Heterodera glycines. J Ultrastr Res 67:1–14
Erb M, Robert CAM (2016) Sequestration of plant secondary metabolites by insect herbivores: molecular mechanisms and ecological consequences. Curr Opin Insect Sci 14:8–11. https://doi.org/10.1016/j.cois.2015.11.005
Erokhina TN, Ryazantsev DY, Zavriev SK, Morozov SY (2023) Regulatory miPEP open reading frames contained in the primary transcripts of microRNAs. Int J Mol Sci 24:2114. https://doi.org/10.3390/ijms24032114
Fanelli E, Dileo C, Di Vito M, De Giorgi C (2008) Inducible antibacterial defence in the plant parasitic nematode Meloidogyne artiellia. Int J Parasitol 38:609–615. https://doi.org/10.1016/j.ijpara.2007.09.002
Félix MA, Ashe A, Piffaretti J, Wu G, Nuez I et al (2011) Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to Nodaviruses. PLoS Biol 9:e1000586. https://doi.org/10.1371/journal.pbio.1000586
Fenn K, Blaxter M (2004) Quantification of Wolbachia bacteria in Brugia malayi through the nematode lifecycle. Mol Biochem Parasitol 137:361–364. https://doi.org/10.1016/j.molbiopara.2004.06.012
Fisher ML, Watson DW, Osborne JA, Mochizuki H, Breen M, Schal C (2018) Growth kinetics of endosymbiont Wolbachia in the common bed bug, Cimex lectularius. Sci Rep 8:11444. https://doi.org/10.1038/s41598-018-29682-2
Flick R, Elgh F, Pettersson RF (2002) Mutational analysis of the Uukuniemi virus (Bunyaviridae family) promoter reveals two elements of functional importance. J Virol 76:10849–10860. https://doi.org/10.1128/jvi.76.21.10849-10860.2002
Flórez LV, Biedermann PHW, Engl T, Kaltenpoth M (2015) Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep 32:904–936. https://doi.org/10.1039/C5NP00010F
Franz CJ, Renshaw H, Frezal L, Jiang Y, Félix MA, Wang D (2014) Orsay, Santeuil and LeBlanc viruses primarily infect intestinal cells in Caenorhabditis nematodes. Virol 448:255–264. https://doi.org/10.1016/j.virol.2013.09.024
Franz CJ, Zhao G, Félix MA, Wang D (2012) Complete genome sequence of Le Blanc virus, a third Caenorhabditis nematode-infecting virus. J Virol 86:11940. https://doi.org/10.1128/JVI.02025-12
Frézal L, Jung H, Tahan S, Wang D, Félix MA (2019) Noda-Like RNA viruses infecting Caenorhabditis nematodes: sympatry, diversity, and reassortment. J Virol 93:e01170-e1219. https://doi.org/10.1128/JVI.01170-19
Fujii C, Wang D (2023) Novel insights into virus-host interactions using the model organism C. elegans. Adv Vir Res 115:135–158. https://doi.org/10.1016/bs.aivir.2023.03.001
Gegner T, Carrau T, Vilcinskas A, Lee KZ (2018) The infection of Harmonia axyridis by a parasitic nematode is mediated by entomopathogenic bacteria and triggers sex-specific host immune responses. Sci Rep 8:15938. https://doi.org/10.1038/s41598-018-34278-x
Gowen S, Davies KG, Pembroke B (2008) Potential use of Pasteuria spp. in the management of plant parasitic nematodes. In: Ciancio A, Mukerji KG (eds) Integrated management and biocontrol of vegetable and grain crops nematodes. Springer, Dordrecht, pp 205–219
Gulcu B (2022) Comparison of powder and liquid forms of antifungal metabolites produced by Xenorhabdus szentirmaii, the symbionts of entomopathogenic nematodes, against gray mold Botrytis cinerea. J Agric Sci Technol 24:457–464
Gulsen SH, Tileklioğlu E, Bode E, Cimen H, Ertabaklar H, Ulug D, Ertug S, Wenski SL, Touray M, Hazir C, Bilecenoglu DK (2022) Antiprotozoal activity of different Xenorhabdus and Photorhabdus bacterial secondary metabolites and identification of bioactive compounds using the easyPACId approach. Sci Rep 12:10779. https://doi.org/10.1038/s41598-022-13722-z
Guo F, Castillo P, Li C, Qing X, Li H (2022) Description of Rotylenchus zhongshanensis sp. nov. (Tylenchomorpha: Hoplolaimidae) and discovery of its endosymbiont Cardinium. J Helminthol 96:e48. https://doi.org/10.1017/S0022149X22000384
Guo YR, Hryc CF, Jakana J, Jiang H, Wang D, Chiu W, Zhong W, Tao YJ (2014) Crystal structure of a nematode-infecting virus. PNAS 111:12781–12786. https://doi.org/10.1073/pnas.1407122111
Haegeman A, Vanholme B, Jacob J, Vandekerckhove TTM, Claeys M, Borgonie G, Gheysen G (2009) An endosymbiotic bacterium in a plant-parasitic nematode member of a new Wolbachia supergroup. Int J Parasitol 39:1045–1054. https://doi.org/10.1016/j.ijpara.2009.01.006
Hagen J, Sarkies P, Selkirk ME (2021) Lentiviral transduction facilitates RNA interference in the nematode parasite Nippostrongylus brasiliensis. PLoS Path 17:e1009286. https://doi.org/10.1371/journal.ppat.1009286
Harrison BD, Robinson DJ (1986) Tobraviruses. In: Van Regenmortel MHV, Fraenkel-Conrat H (eds) The plant viruses. Plenum Press, New York, pp 339–369
Hess RT, Poinar GO Jr (1985) Iridoviruses infecting terrestrial isopods and nematodes. Curr Top Microbiol Immunol 116:49–76. https://doi.org/10.1007/978-3-642-70280-8_4
Hewitt WB, Raski DJ, Goheen AC (1958) Nematode vector of soil-borne fanleaf virus of grapevines. Phytopathol 48:586–595
Hewlett TE, Gerber JF, Smith KS (2004) In vitro culture of Pasteuria penetrans. In: Cook R, Hunt DJ (eds) Proceedings of the Fourth International Congress of Nematology, 8–13 June 2002, Tenerife, Spain. Nematology Monographs and Perspectives 2, Brill, Leiden, NL, pp 175–185
Hoerauf A, Volkmann L, Nissen-Paehle K, Schmetz C, Autenrieth I et al (2000) Targeting of Wolbachia endobacteria in Litomosoides sigmodontis: comparison of tetracyclines with chloramphenicol, macrolides and ciprofloxacin. Trop Med Int Health 5:275–279. https://doi.org/10.1046/j.1365-3156.2000.00544.x
Jacob J, Mitreva M, Vanholme B, Gheysen G (2008) Exploring the transcriptome of the burrowing nematode Radopholus similis. Mol Genet Genom 280:1–17. https://doi.org/10.1007/s00438-008-0340-7
Jones AT, McElroy FD, Brown DJF (1981) Tests for transmission of cherry leaf roll virus using Longidorus, Paralongidorus and **phinema nematodes. Ann Appl Biol 99:143–150. https://doi.org/10.1111/j.1744-7348.1981.tb05141.x
Kallali NS, Ouijja A, Goura K, Laasli SE, Kenfaoui J, Benseddik Y, Blenzar A, Joutei AB, El Jarroudi M, Mokrini F, Lahlali R (2024) From soil to host: discovering the tripartite interactions between entomopathogenic nematodes, symbiotic bacteria and insect pests and related challenges. J Nat Pestic Res 7:100065. https://doi.org/10.1016/j.napere.2023.100065
Kaur R, Shropshire JD, Cross KL, Leigh B, Mansueto AJ, Stewart V, Bordenstein SR, Bordenstein SR (2021) Living in the endosymbiotic world of Wolbachia: a centennial review. Cell Host Micr 29:879–893. https://doi.org/10.1016/j.chom.2021.03.006
Kepenekci I, Hazir S, Lewis EE (2016) Evaluation of entomopathogenic nematodes and the supernatants of the in vitro culture medium of their mutualistic bacteria for the control of the root-knot nematodes Meloidogyne incognita and M. arenaria. Pest Manag Sci 72:327–334. https://doi.org/10.1002/ps.3998
Kikuchi T, Eves-van den Akker S, Jones JT (2017) Genome evolution of plant-parasitic nematodes. Ann Rev Phytopathol 55:333–354. https://doi.org/10.1146/annurev-phyto-080516-035434
Koonin EV, Wolf YI, Nagasaki K, Dolja VV (2008) The big bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat Rev Microbiol 6:925–939. https://doi.org/10.1038/nrmicro2030
Kranse O, Beasley H, Adams S, Pires-daSilva A, Bell C, Lilley CJ, Urwin PE et al (2021) Toward genetic modification of plant-parasitic nematodes: delivery of macromolecules to adults and expression of exogenous mRNA in second stage juveniles. G3-Gene Genom Genet 11:jkaa058. https://doi.org/10.1093/g3journal/jkaa058
Kud J, Dahan J, Orellana GE, Dandurand LM, Karasev AV (2022) A novel Rhabdovirus associated with the Idaho population of potato cyst nematode Globodera pallida. Viruses 14:2718. https://doi.org/10.3390/v14122718
Kumar A, Murthy S, Kapoor A (2017) Evolution of selective-sequencing approaches for virus discovery and virome analysis. Vir Res 239:172–179. https://doi.org/10.1016/j.virusres.2017.06.005
Kurz CL, Ewbank JJ (2000) Caenorhabditis elegans for the study of host-pathogen interactions. Tr Microbiol 8:142–144. https://doi.org/10.1016/s0966-842x(99)01691-1
Kusakabe A, Wang C, Xu Y, Molnár I, Stock SP (2022) Selective toxicity of secondary metabolites from the entomopathogenic bacterium Photorhabdus luminescens sonorensis against selected plant parasitic nematodes of the Tylenchina suborder. Appl Ind Microbiol 10:e02577-e2621. https://doi.org/10.1128/spectrum.02577-21
Labaude S, Griffin CT (2018) Transmission success of entomopathogenic nematodes used in pest control. Insects 9(2):72. https://doi.org/10.3390/insects9020072
Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, Bécard G, Combier JP (2015) Primary transcripts of microRNAs encode regulatory peptides. Nature 520:90–93. https://doi.org/10.1038/nature14346
Le Gall O, Christian P, Fauquet CM, King AM, Knowles NJ et al (2008) Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch Virol 153:715–727. https://doi.org/10.1007/s00705-008-0041-x
Lee MM, Stock SP (2010) A multilocus approach to assessing co-evolutionary relationships between Steinernema spp. (Nematoda: Steinernematidae) and their bacterial symbionts Xenorhabdus spp. (γ-Proteobacteria: Enterobacteriaceae). Syst Parasitol 77:1–12. https://doi.org/10.1007/s11230-010-9256-9
Lin J, Ye R, Thekke-Veetil T, Staton ME, Arelli PR, Bernard EC, Hewezi T et al (2018) A novel picornavirus-like genome from transcriptome sequencing of sugar beet cyst nematode represents a new putative genus. J Gen Vir 99:1418–1424. https://doi.org/10.1099/jgv.0.001139
Jr L, Sullivan T, Schuster ML (1959) A virus disease of Meloidogyne incognita incognita, the southern root knot nematode. Nature 184(Suppl 24):1896. https://doi.org/10.1038/1841896a0
Loulou A, Mastore M, Caramella S, Bhat AH, Brivio MF, Machado RAR et al (2023) Entomopathogenic potential of bacteria associated with soil-borne nematodes and insect immune responses to their infection. PLoS ONE 18(1):e0280675. https://doi.org/10.1371/journal.pone.0280675
Luc JE, Pang W, Crow WT, Giblin-Davis RM (2010) Effects of formulation and host nematode density on the ability of in vitro-produced Pasteuria endospores to control its host Belonolaimus longicaudatus. J Nematol 42:87–90
MacFarlane SA (2003) Molecular determinants of the transmission of plant viruses by nematodes. Mol Plant Pathol 4:211–215. https://doi.org/10.1046/j.1364-3703.2003.00164.x
Maher AMD, Asaiyah MAM, Brophy C, Griffin CT (2017) An entomopathogenic nematode extends its niche by associating with different symbionts. Microb Ecol 73:211–223. https://doi.org/10.1007/s00248-016-0829-2
Machado RAR, Thönen L, Arce CCM, Theepan V, Prada F, Wüthrich D, Robert CAM, Vogiatzaki E, Shi YM, Schaeren OP, Notter M, Bruggmann R, Hapfelmeier S, Bode HB, Erb M (2020) Engineering bacterial symbionts of nematodes improves their biocontrol potential to counter the western corn rootworm. Nat Biotechnol 38:600–608. https://doi.org/10.1038/s41587-020-0419-1
Machado RAR, Bhat AH, Castaneda-Alvarez C, Askary TH, Půža V, Pagès S, Abolafia J (2023a) Xenorhabdus aichiensis sp. nov., Xenorhabdus anantnagensis sp. nov., and Xenorhabdus yunnanensis sp. nov., isolated from Steinernema entomopathogenic nematodes. Curr Microbiol 80:300. https://doi.org/10.1007/s00284-023-03373-2
Machado RAR, Bhat AH, Castaneda-Alvarez C, Půža V, San-Blas E (2023b) Photorhabdus aballayi sp. nov. and Photorhabdus luminescens subsp. venezuelensis subsp. nov., isolated from Heterorhabditis amazonensis entomopathogenic nematodes. Int J Syst Evol Microbiol 73:005872. https://doi.org/10.1099/ijsem.0.005872
Maneesakorn P, An R, Daneshvar H, Taylor K, Bai X, Adams BJ, Grewal PS, Chandrapatya A (2011) Phylogenetic and cophylogenetic relationships of entomopathogenic nematodes (Heterorhabditis: Rhabditida) and their symbiotic bacteria (Photorhabdus: Enterobacteriaceae). Mol Phylogen Evol 59:271–280. https://doi.org/10.1016/j.ympev.2011.02.012
Manoj RRS, Latrofa MS, Epis S, Otranto D (2021) Wolbachia: endosymbiont of onchocercid nematodes and their vectors. Parasit Vect 14:245. https://doi.org/10.1186/s13071-021-04742-1
Martinson VG, Gawryluk RMR, Gowen BE, Curtis CI, Jaenike J, Perlman SJ (2020) Multiple origins of obligate nematode and insect symbionts by a clade of bacteria closely related to plant pathogens. PNAS 117:31979–31986. https://doi.org/10.1073/pnas.2000860117
McMullen JG, Peterson BF, Forst S, Goodrich Blair H, Stock SP (2017) Fitness costs of symbiont switching using entomopathogenic nematodes as a model. BMC Evol Biol 17:100. https://doi.org/10.1186/s12862-017-0939-6
McVeigh P, Alexander-Bowman S, Veal E, Mousley A, Marks NJ, Maule AG (2008) Neuropeptide-like protein diversity in phylum Nematoda. Int J Parasitol 38:1493–1503. https://doi.org/10.1016/j.ijpara.2008.05.006
Mihindukulasuriya KA, Nguyen NL, Wu G, Huang HV, da Rosa AP, Popov VL, Tesh RB, Wang D (2009) Nyamanini and midway viruses define a novel taxon of RNA viruses in the order Mononegavirales. J Virol 83:5109–5116. https://doi.org/10.1128/jvi.02667-08
Moriyama M, Nikoh N, Hosokawa T, Fukatsu T (2015) Riboflavin provisioning underlies Wolbachia’s fitness contribution to its insect host. Mbio 6:e01732-e11715. https://doi.org/10.1128/mBio.01732-15
Murfin KE, Dillman AR, Foster JM, Bulgheresi S, Slatko BE, Sternberg PW, Goodrich-Blair H (2012) Nematode-bacterium symbioses—cooperation and conflict revealed in the “omics” age. Biol Bull 223:85–102. https://doi.org/10.1086/BBLv223n1p85
Nicol JM, Turner SJ, Coyne DL, den Nijs L, Hockland S, Tahna Maafi Z (2011) Current nematode threats to world agriculture. In: Jones J, Gheysen G, Fenoll C (eds) Genomics and molecular genetics of plant-nematode interactions. Springer, Dordrecht, pp 21–44. https://doi.org/10.1007/978-94-007-0434-3
Nikoh N, Hosokawa T, Moriyama M, Oshima K, Hattori M, Fukatsu T (2014) Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc Natl Acad Sci USA 111:10257–10262. https://doi.org/10.1073/pnas.1409284111
Noel G, Atibalentja N (2006) ‘Candidatus Paenicardinium endonii’, an endosymbiont of the plant-parasitic nematode Heterodera glycines (Nemata: Tylenchida), affiliated to the phylum Bacteroidetes. Int J Sys Evol Microbiol 56:1697–1702. https://doi.org/10.1099/ijs.0.64234-0
Ogier JC, Pagès S, Frayssinet M, Gaudriault S (2020) Entomopathogenic nematode-associated microbiota: from monoxenic paradigm to pathobiome. Microbiome 8:25. https://doi.org/10.1186/s40168-020-00800-5
Ogier JC, Akhurst R, Boemare N, Gaudriault S (2023) The endosymbiont and the second bacterial circle of entomopathogenic nematodes. Trends Microbiol 31:629–643. https://doi.org/10.1016/j.tim.2023.01.004
Orlando V, Chitambar JJ, Dong K, Chizhov VN, Mollov D, Bert W, Subbotin SA (2016) Molecular and morphological characterisation of **phinema americanum-group species (Nematoda: Dorylaimida) from California, USA, and other regions, and co-evolution of bacteria from the genus Candidatus **phinematobacter with nematodes. Nematology 18:1015–1043. https://doi.org/10.1163/15685411-00003012
Orr JN, Mauchline TH, Cock PJ, Blok VC, Davies KG (2018) De novo assembly of the Pasteuria penetrans genome reveals high plasticity, host dependency, and BclA-like collagens. bioRxiv. https://doi.org/10.1101/485748
Palomares-Rius JE, Gutiérrez-Gutiérrez C, Mota M, Bert W, Claeys M, Yushin VV, Suzina NE et al (2021) ‘Candidatus **phinematincola pachtaicus’ gen nov., sp nov., an endosymbiotic bacterium associated with nematode species of the genus **phinema (Nematoda, Longidoridae). Int J Syst Evol Microbiol 71:004888. https://doi.org/10.1099/ijsem.0.004888
Parihar RD, Dhiman U, Bhushan A, Gupta PK, Gupta P (2022) Heterorhabditis and Photorhabdus symbiosis: a natural mine of bioactive compounds. Front Microbiol 13:790339. https://doi.org/10.3389/fmicb.2022.790339
Penz T, Schmitz-Esser S, Kelly SE, Cass BN, Muller A, Woyke T, Malfatti SA, Hunter MS, Horn M (2012) Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLOS Gen 8(10):e1003012. https://doi.org/10.1371/journal.pgen.1003012
Pérez-Losada M, Arenas M, Galán JC, Bracho MA, Hillung J, García-González N, González-Candelas F (2020) High-throughput sequencing (HTS) for the analysis of viral populations. Inf Gen Evol 80:104208. https://doi.org/10.1016/j.meegid.2020.104208
Phani V, Somvanshi VS, Shukla RN, Davies KG, Rao U (2018) A transcriptomic snapshot of early molecular communication between Pasteuria penetrans and Meloidogyne incognita. BMC Genom 19:850. https://doi.org/10.1186/s12864-018-5230-8
Poinar GO, Hess-Poinar R (1992) Morphological evidence of a virus infection in the nematode Gastromermis sp. (Mermithidae). J Inv Pathol 60:76–83. https://doi.org/10.1016/0022-2011(92)90157-Y
Poinar GO Jr (1983) The natural history of nematodes. Prentice-Hall Inc, Englewood Cliffs, p 350
Poinar GO Jr (1993) Origins and phylogenetic relationships of the entomophilic rhabditids, Heterorhabditis and Steinernema. Fund Appl Nematol 16:333–338
Porter J, Sullivan W (2023) The cellular lives of Wolbachia. Nature Rev Microbiol 21:750–766. https://doi.org/10.1038/s41579-023-00918-x
Posada-Cespedes S, Seifert D, Beerenwinkel N (2017) Recent advances in inferring viral diversity from high-throughput sequencing data. Vir Res 239:17–32. https://doi.org/10.1016/j.virusres.2016.09.016
Rao RU (2005) Endosymbiotic Wolbachia of parasitic filarial nematodes as drug targets. Indian J Med Res 122:199–204
Raja RK, Arun A, Touray M, Gulsen SH, Cimen H, Gulcu B et al (2021) Antagonists and defense mechanisms of entomopathogenic nematodes and their mutualistic bacteria. Biol Control 152:104452. https://doi.org/10.1016/j.biocontrol.2020.104452
Ritter CL, Malan AP, Dicks LM (2023) Xenorhabdus bakwenae sp. n., associated with the entomopathogenic nematode Steinernema bakwenae. Nematology 25:1169–1179. https://doi.org/10.1163/15685411-bja10284
Robert CAM, Zhang X, Machado RAR, Schirmer S, Lori M, Mateo P, Erb M, Gershenzon J (2017) Sequestration and activation of plant toxins protect the western corn rootworm from enemies at multiple trophic levels. Elife 6:e29307. https://doi.org/10.7554/eLife.29307
Ruark CL, Gardner M, Mitchum MG, Davis EL, Sit TL (2018) Novel RNA viruses within plant parasitic cyst nematodes. PLoS ONE 13(3):e019388. https://doi.org/10.1371/journal.pone.0193881
Ruark CL, Koenning SR, Davis EL, Opperman CH, Lommel SA, Mitchum MG, Sit TL (2017) Soybean cyst nematode culture collections and field populations from North Carolina and Missouri reveal high incidences of infection by viruses. PLoS ONE 12:e0171514. https://doi.org/10.1371/journal.pone.0171514
Sajnaga E, Kazimierczak W (2020) Evolution and taxonomy of nematode-associated entomopathogenic bacteria of the genera Xenorhabdus and Photorhabdus: an overview. Symbiosis 80:1–13. https://doi.org/10.1007/s13199-019-00660-0
Sayedain FS, Ahmadzadeh M, Talaei-Hassanloui R, Olia M, Bode HB (2019) Nematicidal effect of cell-free culture filtrates of EPN- symbiotic bacteria on Meloidogyne javanica. Biol Contr Pests Pl Dis 8:17–26. https://doi.org/10.22059/jbioc.2018.244323.212
Sayre RM, Wergin WP, Schmidt JM, Starr MP (1991) Pasteuria nishizawae sp. nov., a mycelial and endospore forming bacterium parasitic on cyst nematodes of genera Heterodera and Globodera. Res Microbiol 142:551–556
Schellenberger P, Sauter C, Lorber B, Bron P, Trapani S, Bergdoll M, Marmonier A et al (2011) Structural insights into viral determinants of nematode mediated Grapevine fanleaf virus transmission. PLoS Path 7:e1002034. https://doi.org/10.1371/journal.ppat.1002034
Schlesner H, Jenkins C, Staley JT (2006) The phylum Verrucomicrobia: a phylogenetically heterogeneous bacterial group. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes, proteobacteria: delta and epsilon subclasses deeply rooting bacteria, vol 7. Springer. New York, NY, pp 881–896. https://doi.org/10.1007/0-387-30747-8
Shawer R, Donati I, Cellini A, Spinelli F, Mori N (2018) Insecticidal activity of Photorhabdus luminescens against Drosophila suzukii. Insects 9:148. https://doi.org/10.3390/insects9040148
Shepherd AM, Clark SA, Kempton A (1973) An intracellular micro-organism associated with tissues of Heterodera spp. Nematologica 19:31–34
Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC et al (2016) Redefining the invertebrate RNA virosphere. Nature 540:539–543. https://doi.org/10.1038/nature20167
Singh S, Awasthi LP, Jangre A, Nirmalkar VK (2020) Transmission of plant viruses through soil-inhabiting nematode vectors. In: Awasthi LP (ed) Applied plant virology, Chapter 22. Academic Press, Cambridge, pp 291–299. https://doi.org/10.1016/B978-0-12-818654-1.00022-0
Sironi M, Bandi C, Sacchi L, Di Sacco B, Damiani G, Genchi C (1995) Molecular evidence for a close relative of the arthropod endosymbiont Wolbachia in a filarial worm. Mol Biochem Parasitol 74:223–227. https://doi.org/10.1016/0166-6851(95)02494-8
Sivaramakrishnan S, Razia M (2021) Entomopathogenic nematodes and their symbiotic bacteria. Springer protocols handbooks. Springer, New York
Spescha A, Zwyssig M, Hess Hermida M, Moix A, Bruno P, Enkerli J, Campos-Herrera R, Grabenweger G, Maurhofer M (2023) When competitors join forces: consortia of entomopathogenic microorganisms increase killing speed and mortality in leaf- and root-feeding insect hosts. Microb Ecol 86:1947–1960. https://doi.org/10.1007/s00248-023-02191-0
Stackebrandt E (2014) The family Pasteuriaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes. Springer, Berlin, pp 281–284
Stock SP (2019) Partners in crime: symbiont-assisted resource acquisition in Steinernema entomopathogenic nematodes. Curr Opin Insect Sci 32:22–27. https://doi.org/10.1016/j.cois.2018.10.006
Sturhan D, Shutova TS, Akimov VN, Subbotin SA (2005) Occurrence, hosts, morphology, and molecular characterisation of Pasteuria bacteria parasitic in nematodes of the family Plectidae. J Inv Pathol 88:17–26. https://doi.org/10.1016/j.jip.2004.11.001
Tarasco E, Fanelli E, Salvemini C, El-Khoury Y, Troccoli A, Vovlas A, De Luca F (2023) Entomopathogenic nematodes and their symbiotic bacteria: from genes to field uses. Front Insect Sci. https://doi.org/10.3389/finsc.2023.1195254
Tarlachkov SV, Efeykin BD, Castillo P, Evtushenko LI, Subbotin SA (2023) Distribution of bacterial endosymbionts of the Cardinium clade in plant-parasitic nematodes. Int J Mol Sc 24:2905. https://doi.org/10.3390/ijms24032905
Takahashi M, Yunker CE, Clifford CM, Nakano W, Fu**o N, Tanifuji K, Thomas LA (1982) Isolation and characterization of Midway virus: a new tick-borne virus related to Nyamanini. J Med Virol 10:181–193. https://doi.org/10.1002/jmv.1890100304
Tarr DEK (2012) Distribution and characteristics of ABFs, cecropins, nemapores, and lysozymes in nematodes. Dev Comp Immunol 36:502–520. https://doi.org/10.1016/j.dci.2011.09.007
Taylor CE, Robertson WM (1975) Acquisition, retention and transmission of viruses by nematodes. In: Lamberti F, Taylor CE, Seinhorst JW (eds) Nematode vectors of plant viruses. NATO Advanced Study Institutes Series (Series A: Life Sciences), vol 2. Springer, Boston
Thies JA, Petersen AD, Barnes DK (1995) Host suitability of forage grasses and legumes for root-lesion nematode Pratylenchus penetrans. Crop Sci 35:1647–1651. https://doi.org/10.2135/cropsci1995.0011183X003500060022x
Timper P (2009) Population dynamics of Meloidogyne arenaria and Pasteuria penetrans in a long-term crop rotation study. J Nematol 41:291–299
Timper P, Liu C, Davis RF, Wu T (2016) Influence of crop production practices on Pasteuria penetrans and suppression of Meloidogyne incognita. Biol Contr 99:64–71. https://doi.org/10.1016/j.biocontrol.2016.04.013
Tobias NJ, Shi YM, Bode HB (2018) Refining the natural product repertoire in entomopathogenic bacteria. Trends Microbiol 26(10):833–840. https://doi.org/10.1016/j.tim.2018.04.007
Toriyama S, Takahashi M, Sano Y, Shimizu T, Ishihama A (1994) Nucleotide sequence of RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J Gen Virol 75:3569–3579. https://doi.org/10.1099/0022-1317-75-12-3569
Tzortzakakis EA (2023) Survival of **phinema index for six years in stored clay soil from a vineyard. Nematology 25:1193–1195
van Sluijs L, Liu J, Schrama M, van Hamond S, Vromans SPJM et al (2021) Virus infection modulates male sexual behaviour in Caenorhabditis elegans. Mol Ecol 30:6776–6790. https://doi.org/10.1111/mec.16179
Vandekerckhove TTM, Willems A, Gillis M, Coomans A (2000) Occurrence of novel verrucomicrobial species, endosymbiotic and associated with parthenogenesis in **phinema americanum group species (Nematoda, Longidoridae). Int J Sys Evol Microbiol 50:2197–2205. https://doi.org/10.1099/00207713-50-6-2197
Verdejo-Lucas S (1992) Seasonal population fluctuations of Meloidogyne spp. and the Pasteuria penetrans group in kiwi orchards. Pl Dis 76:1275–1279
Vicente-Díez I, Pou A, Campos-Herrera R (2023a) Xenorhabdus- and Photorhabdus-based products: status and future perspective in agriculture. In: Koul O (ed) Development and commercialization of biopesticides. Academic Press, Cambridge, pp 81–101. https://doi.org/10.1016/B978-0-323-95290-3.00012-1
Vicente-Díez I, Pou A, Campos-Herrera R (2023b) The deterrent ability of Xenorhabdus nematophila and Photorhabdus laumondii compounds as a potential novel tool for Lobesia botrana (Lepidoptera: Tortricidae) management. J Invertebr Patholy 198:107911. https://doi.org/10.1016/j.jip.2023.107911
Vicente-Díez I, Moreira X, Pastor V, Vilanova M, Pou A, Campos-Herrera R (2023c) Control of post-harvest gray mold (Botrytis cinerea) on grape (Vitis vinifera) and tomato (Solanum lycopersicum) using volatile organic compounds produced by Xenorhabdus nematophila and Photorhabdus laumondii subsp. laumondii. Biocontrol 68:549–563. https://doi.org/10.1007/s10526-023-10212-7
Vieira P, Nemchinov LG (2019) A novel species of RNA virus associated with root lesion nematode Pratylenchus penetrans. J Gen Virol 100:704–708. https://doi.org/10.1099/jgv.0.001246
Vieira P, Peetz A, Mimee B, Saikai K, Mollov D, MacGuidwin A, Zasada I, Nemchinov LG (2020) Prevalence of the root lesion nematode virus (RLNV1) in populations of Pratylenchus penetrans from North America. J Nematol 52:e2020–e2045. https://doi.org/10.21307/jofnem-2020-045
Vieira P, Subbotin SA, Alkharouf N, Eisenback J, Nemchinov LG (2022) Expanding the RNA virome of nematodes and other soil-inhabiting organisms. Virol Evol 8:1–8. https://doi.org/10.1093/ve/veac019
Wang N, Peng H, Liu S, Huang W, Holgado R, Liu-Clarke J, Peng D (2019) Molecular characterization and functional analysis of two new lysozyme genes from soybean cyst nematode (Heterodera glycines). J Integr Agric 18:2806–2813. https://doi.org/10.1016/S2095-3119(19)62766-8
Wang S, Ahmed I, Li X, Nie J, Guo L (2022) Evidence for a novel partitivirus isolated from the entomopathogenic nematode Steinernema ceratophorum. Arch Virol 167:969–972. https://doi.org/10.1007/s00705-021-05314-5
Wang S, Gergerich RC, Wickizer SL, Kim KS (2002) Localization of transmissible and nontransmissible viruses in the vector nematode **phinema americanum. Phytopathology 92:646–653. https://doi.org/10.1094/PHYTO.2002.92.6.646
Wasala SK, Brown AMV, Kang J, Howe DK, Peetz AB, Zasada IA, Denver DR (2019) Variable abundance and distribution of Wolbachia and Cardinium endosymbionts in plant-parasitic nematode field populations. Front Microbiol 10:964. https://doi.org/10.3389/fmicb.2019.00964
Werren JH (1997) Biology of Wolbachia. Ann Rev Entomol 42:587–609. https://doi.org/10.1146/annurev.ento.42.1.587
Williams SH, Che X, Oleynik A, Garcia JA, Muller D, Zabka TS, Firth C et al (2019) Discovery of two highly divergent negative-sense RNA viruses associated with the parasitic nematode, Capillaria hepatica, in wild Mus musculus from New York City. J Gen Virol 100:1350–1362. https://doi.org/10.1099/jgv.0.001315
Yang D, Chen C, Liu Q, Jian H (2017) Comparative analysis of pre- and post-parasitic transcriptomes and mining pioneer effectors of Heterodera avenae. Cell Biosc 7:11. https://doi.org/10.1186/s13578-017-0138-6
Yin J, Zhu H, **a L, Ding X, Hoffmann T, Hoffmann M et al (2015) A new recombineering system for Photorhabdus and Xenorhabdus. Nucleic Acids Res 43:e36. https://doi.org/10.1093/nar/gku1336
Yüksel E, Yıldırım A, İmren M, Canhilal R, Dababat AA (2023) Xenorhabdus and Photorhabdus bacteria as potential candidates for the control of Culex pipiens L. (Diptera: Culicidae), the principal vector of West Nile Virus and lymphatic filariasis. Pathogens 12:1095. https://doi.org/10.3390/pathogens12091095
Acknowledgements
The authors wish to thank Renée Favre† and Michele Cermola (former IIGB CNR, Naples) for assistance with electron microscopy. PL acknowledges support from Agencia Nacional de Promoción Científica y Tecnológica (Préstamo BID, PICT 2020 N. 1342) and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) (PIP 11220200101685).
Funding
Open access funding provided by IPSP - BARI within the CRUI-CARE Agreement. AC acknowledges funding by MIPAF, project “BIOMED”. Authors declare that no link, affiliation, membership, funding or financial holding affected the objectivity of this study.
Author information
Authors and Affiliations
Contributions
LR and AC: Concept review, data analysis, initial draft and final manuscript writing. PL, MC and IP: topics review and analysis of literature data, critical reading of manuscript and contributions to text. All authors revised and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
AC is a JPS associated editor. All other authors declare no conflict of interest.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Ethical approval
Not applicable.
Additional information
Communicated by Antonio Biondi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rosso, L.C., Lax, P., Ciancio, A. et al. The cryptic microbiota of plant parasitic and entomopathogenic nematodes: diversity, effects on host biology and potential in plant protection. J Pest Sci (2024). https://doi.org/10.1007/s10340-024-01783-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10340-024-01783-0