Abstract
Rationale and objectives
Defacing research MRI brain scans is often a mandatory step. With current defacing software, there are issues with Windows compatibility and researcher doubt regarding the adequacy of preservation of brain voxels in non-T1w scans. To address this, we developed PyFaceWipe, a multiplatform software for multiple MRI contrasts, which was evaluated based on its anonymisation ability and effect on downstream processing.
Materials and methods
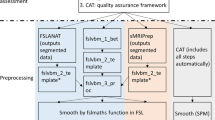
Multiple MRI brain scan contrasts from the OASIS-3 dataset were defaced with PyFaceWipe and PyDeface and manually assessed for brain voxel preservation, remnant facial features and effect on automated face detection. Original and PyFaceWipe-defaced data from locally acquired T1w structural scans underwent volumetry with FastSurfer and brain atlas generation with ANTS.
Results
214 MRI scans of several contrasts from OASIS-3 were successfully processed with both PyFaceWipe and PyDeface. PyFaceWipe maintained complete brain voxel preservation in all tested contrasts except ASL (45%) and DWI (90%), and PyDeface in all tested contrasts except ASL (95%), BOLD (25%), DWI (40%) and T2* (25%). Manual review of PyFaceWipe showed no failures of facial feature removal. Pinna removal was less successful (6% of T1 scans showed residual complete pinna). PyDeface achieved 5.1% failure rate. Automated detection found no faces in PyFaceWipe-defaced scans, 19 faces in PyDeface scans compared with 78 from the 224 original scans. Brain atlas generation showed no significant difference between atlases created from original and defaced data in both young adulthood and late elderly cohorts. Structural volumetry dice scores were ≥ 0.98 for all structures except for grey matter which had 0.93. PyFaceWipe output was identical across the tested operating systems.
Conclusion
PyFaceWipe is a promising multiplatform defacing tool, demonstrating excellent brain voxel preservation and competitive defacing in multiple MRI contrasts, performing favourably against PyDeface. ASL, BOLD, DWI and T2* scans did not produce recognisable 3D renders and hence should not require defacing. Structural volumetry dice scores (≥ 0.98) were higher than previously published FreeSurfer results, except for grey matter which were comparable. The effect is measurable and care should be exercised during studies. ANTS atlas creation showed no significant effect from PyFaceWipe defacing.



Similar content being viewed by others
Data availability
The source data used in this evaluation was from a combination of the OASIS3 database and local clinical scans which we are unable to share. I am happy to share the scripts used to automate generation of the renders and facial recognition. Volumetry data from FastSurfer can be made available on request.
References
Mazura JC, Juluru K, Chen JJ, Morgan TA, John M, Siegel EL (2012) Facial recognition software success rates for the identification of 3D surface reconstructed facial images: implications for patient privacy and security. J Digit Imaging 25(3):347–351. https://doi.org/10.1007/s10278-011-9429-3
Parks CL, Monson KL (2017) Automated facial recognition of computed tomography-derived facial images: patient privacy implications. J Digit Imaging 30(2):204–214. https://doi.org/10.1007/s10278-016-9932-7
Acquisti AGR, Stutzman F (2011) Faces of Facebook: or, how the largest real ID database in the world came to be. Black Hat USA 2011, Las Vegas
Nettrour JF, Burch MB, Bal BS (2019) Patients, pictures, and privacy: managing clinical photographs in the smartphone era. Arthroplast Today 5(1):57–60. https://doi.org/10.1016/j.artd.2018.10.001
Chella F, Marzetti L, Stenroos M, Parkkonen L, Ilmoniemi RJ, Romani GL, Pizzella V (2019) The impact of improved MEG-MRI co-registration on MEG connectivity analysis. Neuroimage 197:354–367. https://doi.org/10.1016/j.neuroimage.2019.04.061
Rubbert C, Wolf L, Turowski B, Hedderich DM, Gaser C, Dahnke R, Caspers J, Alzheimer’s Disease Neuroimaging, I (2022) Impact of defacing on automated brain atrophy estimation. Insights Imaging 13(1):54. https://doi.org/10.1186/s13244-022-01195-7
Schwarz CG, Kremers WK, Wiste HJ, Gunter JL, Vemuri P, Spychalla AJ, Kantarci K, Schultz AP, Sperling RA, Knopman DS, Petersen RC, Jack CR Jr, Alzheimer’s Disease Neuroimaging, I (2021) Changing the face of neuroimaging research: Comparing a new MRI de-facing technique with popular alternatives. Neuroimage 231:117845. https://doi.org/10.1016/j.neuroimage.2021.117845
Theyers AE, Zamyadi M, O’Reilly M, Bartha R, Symons S, MacQueen GM, Hassel S, Lerch JP, Anagnostou E, Lam RW, Frey BN, Milev R, Muller DJ, Kennedy SH, Scott CJM, Strother SC, Arnott SR (2021) Multisite comparison of MRI defacing software across multiple cohorts. Front Psychiatry 12:617997. https://doi.org/10.3389/fpsyt.2021.617997
Cox RW (1996) AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29(3):162–173. https://doi.org/10.1006/cbmr.1996.0014
Khazane A, HJ, Gorgolewksi KJ, Poldrack RA (2019) DeepDefacer: automatic removal of facial features via U-Net image segmentation. ar**v, 2205(15536v1)
Bischoff-Grethe A, Ozyurt IB, Busa E, Quinn BT, Fennema-Notestine C, Clark CP, Morris S, Bondi MW, Jernigan TL, Dale AM, Brown GG, Fischl B (2007) A technique for the deidentification of structural brain MR images. Hum Brain Mapp 28(9):892–903. https://doi.org/10.1002/hbm.20312
Gulban OF, Nielson D, Poldrack R et al (2019) poldracklab/pydeface: v2.0.0. Zenodo. https://doi.org/10.5281/zenodo.3524401
Nakeisha Schimke and John Hale (2011) Quickshear defacing for neuroimages. In: Proceedings of the 2nd USENIX conference on Health security and privacy (HealthSec'11). USENIX Association, USA, 11. https://dl.acm.org/doi/10.5555/2028026.2028037
Mikulan E, Russo S, Zauli FM, d’Orio P, Parmigiani S, Favaro J, Knight W, Squarza S, Perri P, Cardinale F, Avanzini P, Pigorini A (2021) A comparative study between state-of-the-art MRI deidentification and AnonyMI, a new method combining re-identification risk reduction and geometrical preservation. Hum Brain Mapp 42(17):5523–5534. https://doi.org/10.1002/hbm.25639
Milchenko M, Marcus D (2013) Obscuring surface anatomy in volumetric imaging data. Neuroinformatics 11(1):65–75. https://doi.org/10.1007/s12021-012-9160-3
Hoopes A, Mora JS, Dalca AV, Fischl B, Hoffmann M (2022) SynthStrip: skull-strip** for any brain image. Neuroimage 260:119474. https://doi.org/10.1016/j.neuroimage.2022.119474
Fischl B (2012) FreeSurfer. Neuroimage 62(2):774–781. https://doi.org/10.1016/j.neuroimage.2012.01.021
LaMontagne PJ, Benzinger TLS, Morris JC, Keefe S, Hornbeck R, **ong C, Grant E, Hassenstab J, Moulder K, Vlassenko AG, Raichle ME, Cruchaga C, Marcus D (2019) OASIS-3: longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and Alzheimer disease. medRxiv. https://doi.org/10.1101/2019.12.13.19014902
Rosebrock A (2018) Face recognition with OpenCV, Python, and deep learning. Retrieved 15 November from https://pyimagesearch.com/2018/06/18/face-recognition-with-opencv-python-and-deep-learning/. 15 Nov 2023
Deng J, Guo J, Ververas E, Kotsia I, Zafeiriou S (2020) RetinaFace: single-shot multi-level face localisation in the wild. In: 2020 IEEE/CVF Conference on computer vision and pattern recognition (CVPR)
Avants B, Tustison NJ, Song G (2009) Advanced normalization tools: V1.0. Insight J. https://doi.org/10.54294/uvnhin
Gao C, Landman BA, Prince JL, Carass A (2023) A reproducibility evaluation of the effects of MRI defacing on brain segmentation. medRxiv. https://doi.org/10.1101/2023.05.15.23289995
Bhalerao GV, Parekh P, Saini J, Venkatasubramanian G, John JP, consortium, A. (2022) Systematic evaluation of the impact of defacing on quality and volumetric assessments on T1-weighted MR-images. J Neuroradiol 49(3):250–257. https://doi.org/10.1016/j.neurad.2021.03.001
Cali RJ, Bhatt RR, Thomopoulos SI, Gadewar S, Gari IB, Chattopadhyay T, Jahanshad N, Thompson PM (2023) The influence of brain MRI defacing algorithms on brain-age predictions via 3D convolutional neural networks. bioRxiv. https://doi.org/10.1101/2023.04.28.538724
Huo Y, Xu Z, **ong Y, Aboud K, Parvathaneni P, Bao S, Bermudez C, Resnick SM, Cutting LE, Landman BA (2019) 3D whole brain segmentation using spatially localized atlas network tiles. Neuroimage 194:105–119. https://doi.org/10.1016/j.neuroimage.2019.03.041
Acknowledgements
Data were provided in part by OASIS-3 [Longitudinal Multimodal Neuroimaging: Principal Investigators: T. Benzinger, D. Marcus, J. Morris; NIH P30 AG066444, P50 AG00561, P30 NS09857781, P01 AG026276, P01 AG003991, R01 AG043434, UL1 TR000448, R01 EB009352. AV-45 doses were provided by Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical standards
All work was carried out with local ethics board approval.
Conflict of interest
None of the authors have any conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mitew, S., Yeow, L.Y., Ho, C.L. et al. PyFaceWipe: a new defacing tool for almost any MRI contrast. Magn Reson Mater Phy (2024). https://doi.org/10.1007/s10334-024-01170-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10334-024-01170-x