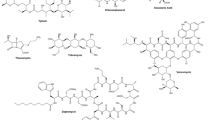
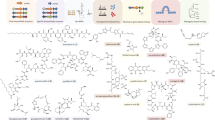
Abstract
Natural product discovery from microorganisms provided important sources for antibiotics, anti-cancer agents, immune-modulators, anthelminthic agents, and insecticides during a span of 50 years starting in the 1940s, then became less productive because of rediscovery issues, low throughput, and lack of relevant new technologies to unveil less abundant or not easily detected drug-like natural products. In the early 2000s, it was observed from genome sequencing that Streptomyces species encode about ten times as many secondary metabolites as predicted from known secondary metabolomes. This gave rise to a new discovery approach—microbial genome mining. As the cost of genome sequencing dropped, the numbers of sequenced bacteria, fungi and archaea expanded dramatically, and bioinformatic methods were developed to rapidly scan whole genomes for the numbers, types, and novelty of secondary metabolite biosynthetic gene clusters. This methodology enabled the identification of microbial taxa gifted for the biosynthesis of drug-like secondary metabolites. As genome sequencing technology progressed, the realities relevant to drug discovery have emerged, the conjectures and misconceptions have been clarified, and opportunities to reinvigorate microbial drug discovery have crystallized. This perspective addresses these critical issues for drug discovery.



Similar content being viewed by others
References
Adamek M, Alanjary M, Sales-Ortells H et al (2018) Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis. BMC Genom 19:426
Ahlert J, Shepard E, Lomovskaya N et al (2002) The calicheamicin gene cluster and its iterative type I enediyne PKS. Science 297:1173–1176
Aigle B, Lautra S, Spiteller D, Dickshat JS, Challis GL, Leblond P, Pernodet JL (2014) Genome mining in Streptomyces ambofaciens. J Ind Microbiol Biotechnol 41:251–263
Alexander DC, Rock J, He X, Brian P, Miao V, Baltz RH (2010) Development of a genetic system for combinatorial biosynthesis of lipopeptides in Streptomyces fradiae and heterologous expression of the A54145 biosynthetic gene cluster. Appl Environ Microbiol 76:6877–6887
Amann RI, Ludwig W, Scheifer KH (2009) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Bachmann BO, Van Lanen SG, Baltz RH (2014) Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J Ind Microbiol Biotechnol 41:175–184
Bai L, Li L, Xu H, Minagawa K, Yu Y, Zhang Y, Zhou X, Floss HG, Mahmud T, Deng Z (2006) Functional analysis of the validamycin biosynthetic gene cluster and engineered production of validoxylamine A. Chem Biol 13:387–397
Baltz RH (1982) Genetics and biochemistry of tylosin production: a model for genetic engineering in antibiotic-producing Streptomyces. Basic Life Sci 19:431–444
Baltz R (2005) Natural product discovery and development at Eli Lilly and Company: one scientist’s view. SIM News 55:5–16
Baltz RH (2005) Antibiotic discovery from actinomycetes: will a renaissance follow the decline and fall? SIM News 56:148–160
Baltz RH (2006) Molecular engineering approaches to peptide, polyketide and other antibiotics. Nat Biotechnol 24:1533–1540
Baltz RH (2006) Combinatorial biosynthesis of novel antibiotics. SIM News 56:148–160
Baltz RH (2006) Marcel faber roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration. J Ind Microbiol Biotechnol 33:507–513
Baltz RH (2007) Antimicrobials from actinomycetes: back to the future. Microbe 2:125–131
Baltz RH (2008) Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol 8:557–563
Baltz RH, Monahan C, Murphy C, Penn J, Ritz D, Wrigley S (2010) Ultra-high throughput screening of natural products. US Patent Application Pub. No.: US 2010/0311107 A1
Baltz RH (2014) MbtH homology codes to identify gifted microbes for genome mining. J Ind Microbiol Biotechnol 41:357–369
Baltz RH (2014) Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways. ACS Synth Biol 3:748–759
Baltz RH (2016) Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol 43:343–370
Baltz RH (2017) Gifted microbes for genome mining and natural product discovery. J Ind Microbiol Biotechnol 44:573–588
Baltz RH (2017) Molecular beacons to identify gifted microbes for genome mining. J Antibiot 70:639–646
Baltz RH (2017) Microbial genome mining for natural product drug discovery. In: Newman DJ, Cragg GM, Grothaus PG (eds) Chemical biology of natural products. CRC Press, Boca Raton, pp 1–42
Baltz RH (2018) Synthetic biology, genome mining, and combinatorial biosynthesis of NRPS-derived antibiotics: a perspective. J Ind Microbiol Biotechnol 45:635–649
Ban YH, Park SR, Yoon YJ (2016) The biosynthetic pathway of FK506 and its engineering: from past achievements to future prospects. J Ind Microbiol Biotechnol 43:389–400
Barka EA, Vasta P, Sanchez L et al (2016) Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev 80:1–43
Bentley SD, Chater KF, Cwedeno-Tarraga AM et al (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bills GF, Glover JB (2016) Biologically active secondary metabolites from the fungi. Microbiol Spectr 4:FUNK-0009-2016
Bindman NA, Van Der Donk WA (2014) RiPPs: ribosomally synthesized and posttranslationally modified peptides. In: Osbourn A, Goss RJ, Carter G (eds) Natural products: discourse, discovery, and design. Wiley Blackwell, Ames, Iowa, pp 197–217
Blin K, Wolf T, Chevrette MG et al (2017) antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–W41
Boakes S, Dawson MJ (2014) Discovery and development of NVB302, a semisynthetic antibiotic for treatment of Clostridium difficile infection. In: Osbourn A, Goss RJ, Carter G (eds) Natural products: discourse, discovery, and design. Wiley Blackwell, Ames, Iowa, pp 455–468
Braesel J, Crnkovic CM, Kunstman KJ, Green SJ, Maiencheing-Cline M, Orjala J, Murphy BT, Estáquio AS (2018) Complete genome of Micromonospora sp. strain B006 reveals biosynthetic potential of a lake Michigan actinomycete. J Nat Prod 81:2057–2068
Brautaset T, Sekurova ON, Stetta H, Ellingsen TE, Strøm AR, Valla S, Zotchev SB (2000) Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem Biol 7:395–403
Bull AT, Idris H, Sanderson R, Asenjo J, Andrews B, Goodfellow M (2018) High altitude, hyper-arid soils of the Central-Andes harbor mega-diverse communities of actinobacteria. Extremophiles 22:47–57
Ceniceros A, Dijkhuizen L, Petrusma M, Medema MH (2017) Genome-based exploration of the specialized metabolic capabilities of the genus Rhodococcus. BMC Genom 18:593
Challis GL (2014) Exploitation of the Streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J Ind Microbiol Biotechnol 41:219–232
Charlop-Powers Z, Owen JG, Reddy BVB et al (2015) Global biogeographic sampling of bacterial secondary metabolism. eLife 4:e05048
Chen S, Huang X, Zhou X, Bai L, He J, Jeong KJ, Lee SY, Deng Z (2003) Organization and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem Biol 10:1065–1076
Chen W, Qu D, Zhai L, Tao M, Wang Y, Lin S, Price NP, Deng Z (2010) Characterization of the tunicamycin gene cluster unveiling unique steps involved in its biosynthesis. Protein Cell 1:1093–1105
Chiang YM, Wang CCC, Oakley BR (2014) Analyzing fungal secondary metabolite genes and gene clusters. In: Osbourn A, Goss RJ, Carter G (eds) Natural products: discourse, discovery, and design. Wiley Blackwell, Ames, Iowa, pp 103–124
Cimermancic P, Medema MH, Claesen J et al (2014) Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158:412–421
Cox RJ, Williams K (2014) Manipulation of fungal natural product pathways. In: Osbourn A, Goss RJ, Carter G (eds) Natural products: discourse, discovery, and design. Wiley Blackwell, Ames, Iowa, pp 245–260
Cundliffe E (2008) Control of tylosin biosynthesis in Streptomyces fradiae. J Microbiol Biotechnol 18:1485–1491
Demain AL (2014) Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biotechnol 41:185–201
Donia MS, Cimermancic P, Schulze CJ et al (2014) A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158:1402–1414
Donia MS, Fischbach MA (2015) HUMAN MICROBIOTA: small molecules from the human microbiota. Science 349:1254766
Fenagle EA, Rondon MR, Berti AD, Crosby HA, Thomas MG (2007) Identification of the biosynthetic gene cluster and an additional gene for resistance to the antituberculosis drug capreomycin. Appl Environ Microbiol 73:4162–4170
Genilloud O, González L, Salizar O, Martin J, Tormo JR, Vincente F (2011) Current approaches to exploit actinomycetes as a source of novel natural products. J Ind Microbiol Biotechnol 38:375–389
Gomez-Escribano JP, Castro JF, Razmilic V, Chandra G, Andrews B, Bibb MJ (2016) The Streptomyces leeuwenhoekii genome: de novo sequencing and assembly in single contigs of the chromosome, circular plasmid pSLE1 and linear plasmid pSLE2. BMC Genom 16:485
Gomez-Escribano JP, Alt S, Bibb MJ (2016) Next generation sequencing of actinobacteria for the discovery of novel natural products. Mar Drugs 14:78
Goodfellow M, Nouioui I, Sanderson R, **e F, Bull AT (2018) Rare taxa and dark microbial matter: novel bioactive actinobacteria abound in Atacama Desert soils. Antonie Van Leeuwenhoek 111:1315–1332
Gu W, Schmidt EW (2017) Three principles of diversity-generating biosynthesis. Acc Chem Res 50:569–2576
Gulick AM, Aldrich CC (2018) Trap** interactions between catalytic domains and carrier proteins of modular biosynthetic enzymes with chemical probes. Nat Prod Rep 35:1156–1184
Gullo VP, McAlpine J, Lam KS, Baker D, Petersen F (2006) Drug discovery from natural products. J Ind Microbiol Biotechnol 33:523–531
Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5:R245–R249
He Y, Wang B, Chen W, Cox RJ, He J, Chen F (2018) Recent advances in reconstructing microbial secondary metabolites biosynthesis in Aspergillus spp. Biotechnol Adv 36:739–783
Hoffmann T, Krug D, Bozkurt N et al (2018) Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat Commun 9:803
Iftime D, Kulik A, Härtner T et al (2016) Identification and activation of novel biosynthetic gene clusters by genome mining in the kirromycin producer Streptomyces collinus Tu 365. J Ind Microbiol Biotechnol 43:277–291
Ikeda H, Nonomita T, Usami M, Ohta T, Ōmura S (1999) Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc Nat Acad Sci USA 96:9509–9514
Ikeda H, Ishikawa J, Hanamoto A et al (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531
Ikeda H, Shin-ya K, Ōmura S (2014) Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol 41:233–250
Karray F, Darbon E, Oestreicher N et al (2007) Organization of the biosynthetic gene cluster for the macrolide antibiotic spiramycin in Streptomyces ambofaciens. Microbiology 153:4111–4122
Karwehl S, Stadler M (2016) Exploration of fungal biodiversity for discovery of novel antibiotics. Curr Top Microbiol Immunol 398:303–338
Katz L (2009) The DEBS paradigm for type I modular polyketide synthases and beyond. Methods Enzymol 459:113–142
Katz L, Baltz RH (2016) Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol 43:155–176
Katz L, Chen YY, Gonzalez R, Peterson TC, Zhao H, Baltz RH (2018) Synthetic biology advances and applications in the biotechnology industry: a perspective. J Ind Microbiol Biotechnol 45:449–461
Kharel MK, Subba B, Basnet DB, Woo JS, Lee HC, Liou K, Sohng JK (2004) A gene cluster for biosynthesis of kanamycin from Streptomyces kanamyceticus: comparison with gentimicin biosynthetic gene cluster. Arch Biochem Biophys 429:204–214
Kim KR, Kim TJ, Suh JW (2008) The gene cluster for spectinomycin biosynthesis and the aminoglycoside-resistance function of spcM in Streptomyces spectabilis. Curr Microbiol 57:371–374
Koběrská M, Kopecky J, Olšovská J et al (2008) Sequence analysis and heterologous expression of the lincomycin biosynthetic gene cluster of the type strain Streptomyces lincolnensis ATCC 25466. Folia Microbiol 53:395–401
Kouzes JM, Posner BZ (1995) The leadership challenge, 2nd edn. Jossey-Bass, San Francisco
Kudo F, Miyanaga A, Eguchi T (2018) Structural basis of the nonribosomal codes for nonproteinogenic amino acid selective adenylation enzymes in the biosynthesis of natural products. J Ind Microbiol Biotechnol. https://doi.org/10.1007/s10295-018-2084-7
Li YF, Tsai KJ, Harvey CJ et al (2016) Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products. Fungal Genet Biol 89:18–28
Liao G, Li J, Li L, Tian Y, Tan H (2010) Cloning, reassembling and integration of the entire nikkomycin biosynthetic gene cluster into Streptomyces ansochromogenes lead to an improved nikkomycin production. Microb Cell Fact 9:6
Liao R, Duan L, Lei C et al (2009) Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol 16:141–147
Lim YH, Wong FT, Yeo WL et al (2018) Auroramycin: a potent antibiotic from Streptomyces roseosporus by CRISPR–Cas9 activation. Chembiochem. https://doi.org/10.1002/cbic.201800266
Liu X, Cheng YQ (2014) Genome-guided discovery of diverse natural products from Burkholderia sp. J Ind Microbiol Biotechnol 41:275–284
Lu C, Zhang X, Jiang M, Bai L (2016) Enhanced salinomycin production by adjusting the supply of polyketide extender units in Streptomyces albus. Metab Eng 35:129–137
Mahler L, Wink K, Beulig RJ et al (2018) Detection of antibiotics synthesized in microfluidic picolitre-droplets by various actinobactereia. Sci Rep 8:13087
Mao Y, Varaglu M, Sherman DH (1999) Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chem Biol 6:251–263
Marcone GL, Binda E, Berini F, Marinelli F (2018) Old and new glycopeptide antibiotics: from product to gene and back in the post-genomic era. Biotechnol Adv 36:534–554
McDonald BR, Currie CR (2017) Lateral transfer dynamics in the ancient bacterial genus Streptomyces. mBio 8:e00644-17
Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA (2014) A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput Biol 10:e1004016
Medema MH, Kottmann R, Yilmaz P et al (2015) Minimum information about a biosynthetic gene cluster. Nat Chem Biol 11:625–631
Miao V, Coëffet-Le Gal M-F, Brian P et al (2005) Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151:1505–1523
Miao V, Brost R, Chapple J, She K, Coëffet-Le Gal MF, Baltz RH (2006) The lipopeptide antibiotic A54145 gene cluster from Streptomyces fradiae. J Ind Microbiol Biotechnol 33:129–140
Mukherjee S, Seshadri R, Varghese NJ et al (2017) 1003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nat Biotechnol 35:676–683
Müller C, Nolden S, Gebhardt P, Heinzelmann E, Lange C, Puk O, Welzel K, Wohlleben W, Schwartz D (2007) Sequencing and analysis of the biosynthetic gene cluster of the lipopeptide antibiotic friulimicin in Actinoplanes friuliensis. Antimicrob Agents Chemother 51:1028–1037
Musiol-Kroll EM, Wohlleben W (2018) Acyltransferases as tools for polyketide synthase engineering. Antibiotics 7:E62
Nakano H, Ōmura S (2009) Chemical biology of natural indolcarbazole products: 30 years since the discovery of staurosporine. J Antibiot 62:17–26
Nett M, Ikeda H, Moore B (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26:1362–1384
Newman DJ, Cragg GM (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79:629–661
Novakova R, Núnez LE, Homorova D et al (2017) Increased heterologous production of the antitumor polyketide mithramycin A by engineered Streptomyces lividans TK24 strains. Appl Microobiol Biotechnol 102:857–869
Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S (2008) Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol 190:4050–4060
Olano C, Mendez C, Salas JA (2014) Harnessing sugar biosynthesis and glycosylation to redesign natural products and to increase structural diversity. In: Osbourn A, Goss RJ, Carter G (eds) Natural products: discourse, discovery, and design. Wiley Blackwell, Ames, Iowa, pp 317–339
Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25:447–453
Pace NR (2009) Map** the tree of life. Microbiol Mol Biol Rev 73:565–576
Pantel L, Florin T, Dobosz-Bartoszek M et al (2018) Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. Mol Cell 70:83–94
Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL (2007) Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40
Petrović H, Cullum J, Hranuelii D et al (2006) genetics of Streptomyces rimosus, the oxytetracycline producer. Microbiol Mol Biol Rev 70:704–728
Plaza A, Müller R (2014) Myxobacteria: chemical diversity and screening strategies. In: Osbourn A, Goss RJ, Carter G (eds) Natural products: discourse, discovery, and design. Wiley Blackwell, Ames, Iowa, pp 103–124
Pojer F, Li SM, Heide L (2002) Molecular cloning and sequence analysis of the chlorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901–3911
Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG (2017) Retrospective analysis of natural products provides insights for future discovery trends. Proc Nat Acad Sci USA 114:5601–5606
Reimer JM, Haque AS, Tarry MJ, Schmeing TM (2018) Piecing together nonribosomal peptide synthesis. Curr Opin Struct Biol 49:104–113
Ren H, Wang B, Zhao H (2017) Breaking the silence: new strategies for discovering novel natural products. Curr Opin Biotechnol 48:21–27
Schwalen CJ, Hudson GA, Kille B, Mitchell DA (2018) Bioinformatic expansion and discovery of thiopeptide antibiotics. J Am Chem Soc 140:9494–9501
Schwartz D, Berger S, Heinzelmann E, Muschko K, Welzel K, Wohlleben W (2004) Biosynthetic gene cluster of the herbicide phosphinothricin tripeptide from Streptomyces viridochromogenes Tü94. Appl Environ Microbiol 70:7093–7102
Schwecke T, Aparicio JF, Molnar I et al (1995) The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Nat Acad Sci USA 92:7839–7843
Seipke RF (2015) Strain-level diversity of secondary metabolites in Streptomyces albus. PLoS One 10:e0116457
Senges CH, Al-Dilaimi A, Marchbank DH, Wibberg D, Winkler A, Haltli B, Nowrousian M, Kalinowski J, Kerr R, Bandow JE (2018) The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc Nat Acad Sci USA 115:2490–2495
Shetty SA, Hugenholtz F, Lahti L, Smidt H, de Vos WM (2017) Intestinal microbiome landsca**: insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol Rev 41:182–199
Smanski MJ, Casper J, Peterson RM, Rajski SR, Shen B (2012) Expression of the platencin biosynthetic gene cluster in heterologous hosts yielding new platencin congeners. J Nat Prod 75:2158–2167
Smanski MJ, Schlatter DC, Kinkel LL (2016) Leveraging ecological theory to guide natural product discovery. J Ind Microbiol Biotechnol 43:115–128
Smith L, Hong H, Spencer JB, Leadlay PF (2008) Analysis of specific mutants in the lasalocid gene cluster: evidence for enzymatic catalysis of a disfavored polyether ring closure. ChemBioChem 9:2967–2975
Spraker J, Keller N (2014) Waking slee** pathways in filamentous fungi. In: Osbourn A, Goss RJ, Carter G (eds) Natural products: discourse, discovery, and design. Wiley Blackwell, Ames, Iowa, pp 279–292
Süssmuth RD, Mainz A (2017) Nonribosomal peptide synthesis-principle and prospects. Angew Chem Int Ed 56:3770–3823
Tao W, Yang A, Deng Z, Sun Y (2018) CRISPR/Cas9-based editing of Streptomyces for discovery, characterization, and production of natural products. Front Microbiol 9:1660. https://doi.org/10.3389/fmicb.2018.01660
Tietz JI, Patel PS, Maxson T, Blair PM, Tai HC, Zakai UI, Mitchell DA (2017) A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol 13:470–478
Tobias NJ, Wolff H, Djahanschiri B et al (2017) Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat Microbiol 2:1676–1685
Van der Lee TA, Medema MH (2016) Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Genet Biol 89:29–36
Waldron C, Matsushima P, Rosteck PR, Broughton MC, Turner J, Madduri K, Crawford KP, Merlo DJ, Baltz RH (2001) Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem Biol 8:487–499
Wang XJ, Yan YJ, Zhang B et al (2010) Genome sequence of the milbamycin-producing Streptomyces bingchenggensis. J Bacteriol 192:4526–4527
Wang Y, Chen Y, Shen Q, Yin X (2011) Molecular cloning and identification of the laspartomycin biosynthetic gene cluster from Streptomyces viridochromogenes. Gene 483:11–21
Weber T, Blin K, Duddela S et al (2015) antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243
Weissman KJ (2015) The structural biology of biosynthetic megaenzymes. Nat Chem Biol 11:660–670
Wiemann P, Keller NP (2014) Strategies for mining fungal natural products. J Ind Microbiol Biotechnol 41:301–313
**ao Y, Li S, Niu S, Ma L, Zhang G, Zhang H, Ju J, Zhang C (2011) Characterization oftiacumicin B biosynthetic gene cluster affording diversified tiacumicin analogs and revealing a tailoring dihalogenase. J Am Chem Soc 133:1092–1105
Xu M, Wright GD (2018) Heterologous expression-facilitated natural products discovery in actinomycetes. J Ind Microbiol Biotechnol. https://doi.org/10.1007/s10295-018-2097-2
Yaegashi J, Oakley BR, Wang CC (2014) Recent advances in genome mining fungal natural products. J Ind Microbiol Biotechnol 41:433–442
Yuzawa S, Backman TW, Keasling JD, Katz L (2018) Synthetic biology of polyketide synthases. J Ind Microbiol Biotechnol 45:621–633
Zhang MM, Wong FT, Wang Y, Luo S, Lim YH, Heng E, Yeo WK, Cobb RE, Enghiad B, Ang E, Zhao H (2017) CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol 13:607–609
Zhao W, Zhong Y, Yuan H et al (2010) Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res 20:1096–1108
Acknowledgements
I thank Heinz Floss and Chris Walsh for their many contributions to the field on natural product biosynthesis. I thank all contributors to this Special Issue of JIMB devoted to natural products. It is my hope that advances in natural product research continue at an accelerated pace, and contribute to a resurgence in natural product discovery, particularly for antimicrobials active against drug-resistant pathogens. As a father and grandfather, I look forward to the “Platinum Age” of natural product drug discovery for current and future generations.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Special Issue “Natural Product Discovery and Development in the Genomic Era 2019”
Rights and permissions
About this article
Cite this article
Baltz, R.H. Natural product drug discovery in the genomic era: realities, conjectures, misconceptions, and opportunities. J Ind Microbiol Biotechnol 46, 281–299 (2019). https://doi.org/10.1007/s10295-018-2115-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2115-4