Abstract
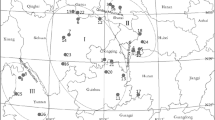
Amentotaxus, a genus of the Taxaceae, represents an ancient lineage that has long existed in Eurasia. All Amentotaxus species experienced frequent population expansion and contraction over periodical glaciations in Tertiary and Quaternary. Among them, Amentotaxus argotaenia complex consists of three morphologically alike species, A. argotaenia, Amentotaxus yunnanensis, and Amentotaxus formosana. This complex is distributed in the subtropical region of mainland China and Taiwan where many Pleistocene refugia have been documented. In this study, genetic diversity and population structuring within and between species were investigated based on the inter-simple sequence repeats (ISSR) fingerprinting. Mean genetic diversity within populations was estimated in three ways: (1) the percentage of polymorphic loci out of all loci (P) (2) Nei’s unbiased expected heterozygosity (He), and (3) Shannon’s index of phenotypic diversity. For a total of 310 individuals of 15 populations sampled from the three species, low levels of ISSR genetic variation within populations were detected, with P=4.66–16.58%, He=0.0176–0.0645 and Hpop=0.0263–0.0939, agreeing with their seriously threatened status. AMOVA analyses revealed that the differences between species only accounted for 27.38% of the total variation, whereas differences among populations and within populations were 57.70 and 14.92%, respectively, indicating substantial isolation between the patch-like populations. A neighbor-joining tree identified a close affinity between A. yunnanensis and A. formosana. Genetic drift due to small population size, plus limited current gene flow, resulted in significant genetic structuring. Low levels of intrapopulational genetic variation and considerable interpopulational divergence were also attributable to demographic bottlenecks during and/or after the Pleistocene glaciations.


Similar content being viewed by others
References
Ash GJ, Raman R, Crump NS (2003) An investigation of genetic variation in Carthamus lanatus in New South Wales, Australia, using intersimple sequence repeats (ISSR) analysis. Weed Res 43:208–213
Axelrod DI, Al-Shehbaz I, Raven PH (1998) History of the modern flora of China. In: Zhang AL, Wu SG (eds) Floristic characteristics and diversity of East Asian plants. China Higher Education Press/Springer, Bei**g/Berlin Heidelberg New York, pp 43–55
Bauert MR, Kalin M, Baltisberger M, Edwards PJ (1998) No genetic variation detected within isolated relict populations of Saxifraga cernua in the Alps using RAPD markers. Mol Ecol 7:1519–1527
Cheng WC (1978) The genus Amentotaxus. Flora Reipublicae Popularis Sinicae, vol 7. Science Press, Bei**g
Chiang TY, Hong KH, Peng CI (2001) Experimental hybridization reveals biased inheritance of the internal transcribed spacer of the nuclear ribosomal DNA in Begonia×taipeiensis. J Plant Res 114:343–351
Chiang YC, Schaal BA, Chou CH, Huang S, Chiang TY (2003) Contrasting selection modes at the Adh 1 1 locus in outcrossing Miscanthus sinensis vs inbreeding Miscanthus condensatus (Poaceae). Am J Bot 90:561–570
Comes HP, Kadereit JW (1998) The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci 3:432–438
Culley TM, Wolfe AD (2001) Population genetic structure of the cleistogamous plant species Viola pubescens Aiton (Violaceae), as indicated by allozyme and ISSR molecular markers. Heredity 86:545–556
Doyle J (1991) DNA protocols for plants—CTAB total DNA isolation. In: Hewitt GM, Johnston A (eds) Molecular techniques in taxonomy. Springer, Berlin Heidelberg New York, pp 283–293
Dyer RJ, Sork VL (2001) Pollen pool heterogeneity in shortleaf pine, Pinus echinata Mill. Mol Ecol 10:859–866
Farjon A (2001) World checklist and bibliography of conifers, 2nd edn. The Royal Botanic Gardens Press, Kew, England
Felsenstein J (1993) PHYLIP, Phylogenetic inference package, version 3.5.7. A computer program distributed by the author, http://evolution. genetics.washington.edu/phylip.html. Department of genetics, University of Washington, Seattle
Ferguson DK, Jahnichen H, Alvin KL (1978) Amentotaxus Pilger from the European tertiary. Feddes Rep 89:379–410
Fontaine C, Lovett PN, Sanou H, Maley J, Bouvet J-M (2004) Genetic diversity of the shea tree (Vitellaria paradoxa C.F. Gaertn), detected by RAPD and chloroplast microsatellite markers. Heredity 93:639–648
Frankel OH, Soulé ME (1981) Conservation and evolution. Cambridge University Press, Cambridge
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Gadek PA, Alpers DL, Heslewood MM, Quinn CJ (2000) Relationships within Cupressaceae sensu lato: a combined morphological and molecular approach. Am J Bot 87:1044–1057
Ge S, Hong DY, Wang HQ, Liu ZY, Zhang CM (1998) Population genetic structure and conservation of an endangered conifer, Cathaya argyrophylla (Pinaceae). Int J Plant Sci 159:351–357
Ge XJ, Yu Y, Zhao NX, Chen HS, Qi WQ (2003) Genetic variation in the endangered inner Mongolia endemic shrub Tetraena mongolica Maxim. (Zygophyllaceae). Biol Conserv 111:427–434
Ge XJ, Yu Y, Yuan YM, Huang HW, Yang C (2005) Genetic diversity and geographic differentiation in endangered Ammopiptanthus (Leguminosae) populations in desert regions of Northwest China as revealed by ISSR analysis. Ann Bot 95:843–851
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding and genetic resources. Sinauer Associates, Sunderland, MA, pp 43–63
Hamrick JL, Godt MJW (1996) Conservation genetics of endemic plant species. In: Avise JC, Hamrick JL (eds) Conservation genetics: case histories from nature. Chapman and Hall, New York, pp 281–304
Hamrick JL, Godt MJW, Murawski DA, Loveless MD (1991) Correlations between species and allozyme diversity: implications for conservation biology. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 75–86
Hartl DL, Clark AG (1997) Principles of population genetics, 3rd edn. Sinauer, Sunderland, MA
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Holsinger KE, Lewis PO, Dey DK (2002) A Bayesian approach to inferring population structure from dominant markers. Mol Ecol 11:1157–1164
Karron JD (1991) Patterns of genetic variation and breeding systems in rare plants. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 87–98
Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:381–398
Lin TP, Lee TY, Yang LF, Chung YL, Yang JC (1994) Comparison of the allozyme diversity in several populations of Chamaecyparis formosensis and Chamaecyparis taiwanensis. Can J Forest Res 24:2128–2134
Lu SY, Peng CI, Cheng YP, Hong KH, Chiang TY (2001) Chloroplast DNA phylogeography of Cunninghamia konishii (Cupressaceae), an endemic conifer of Taiwan. Genome 44:797–807
Maki M (2003) Population genetics of threatened wild plants in Japan. J Plant Res 116:169–174
Miller CN Jr (1977) Mesozoic conifers. Bot Rev 43:217–280
Miller MP (1997) Tools for population genetic analysis (TFPGA), version 1.3. Department of Biological Sciences, Northern Arizona University, AZ
Nei M (1972) Genetic distance between populations. Am Nat 106:282–292
Nichols RA, Hewitt GM (1994) The genetic consequences of long distance dispersal during colonization. Heredity 72:312–317
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol Evol Syst 3(2):93–114
Page CN (1990) Cupressaceae. In: Kubitzky K (eds) The families and genera of vascular plants vol. 1. Springer, Berlin Heidelberg New York, pp 302–316
Royer DL, Hickey LJ, Wing SL (2003) Ecological conservatism in the “living fossil” Ginkgo. Paleobiology 29:84–104
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva
Sibuet JC, Hsu SK (1997) Geodynamics of the Taiwan arc–arc collision. Tectonophysics 274:221–251
Sibuet JC, Hsu SK (2004) How was Taiwan created? Tectonophysics 279:159–181
Suzuki H, Tsuchiya K, Takezaki N (2000) A molecular phylogenetic framework for the Ryukyu endemic rodents Tokudaia osimensis and Diplothrix legata. Mol Phylogen Evol 15:15–24
Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Templeton AR, Routman E, Phillips CA (1995) Separating population structure from population history: a cladistic analysis the geographical distribution of mitochondrial DNA haplotypes in the tiger salamander, Ambystoma tigrinum. Genetics 140:767–782
Walter R, Epperson BK (2004) Microsatellite analysis of spatial structure among seedlings in populations of Pinus strobus (Pinaceae). Am J Bot 91:549–557
Wang WT (1992) On some distribution patterns and some migration routes found in the Eastern Asiatic region. Acta Phytotax Sin 30 1–24
Wang CT, Wang WT, Chiang CH, Wang YN, Lin TP (1996) Low genetic variation in Amentotaxus formosana Li revealed by isozyme analysis and random amplified polymorphic DNA markers. Heredity 77:388–395
Wang T, Su YJ, Li XY, Chen GP, Zeng QL (2004) Genetic structure and variation in the relict populations of Alsophila spinulosa from southern China based on RAPD markers and cpDNA atpB-rbcL sequence data. Hereditas 140:8–17
**ao LQ, Ge XJ, Gong X, Hao G, Zhang SX (2004) ISSR variation in the endemic and endangered plant Cycas guizhouensis (Cycadaceae). Ann Bot 94:133–138
**e GW, Wang SL, Yuan YM, Ge XJ (2005) Population genetic structure of Monimopetalum chinense (Celastraceae). Ann Bot 95:773–777
Yeh FC, Yang RC, Boyle T (1999) POPGENE. Microsoft windows-based freeware for population genetic analysis. Release 1.31. University of Alberta, Edmonton, AB
Zeng WB (1993) The passageway of the flora migration on both sides of the Taiwan Strait in Pleistocene epoch. Acta Bot Yunn 16:107–110
Zhou QX (2001) Studies on Systematics of Taxaceae. PhD Dissertation. Kunming Institute of Botany, Kunming, pp 1–133
Zietkiewicz E, Rafalski A, Labud D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
We thank Chao-Yi Deng, Guo-Sheng He, Yun-Fei Deng, Hua-Gu Ye, and Qing-Zhou Zheng for their help with material sampling. This work was supported by the National Natural Science Foundation of China (No. 30170070).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ge, XJ., Zhou, XL., Li, ZC. et al. Low genetic diversity and significant population structuring in the relict Amentotaxus argotaenia complex (Taxaceae) based on ISSR fingerprinting. J Plant Res 118, 415–422 (2005). https://doi.org/10.1007/s10265-005-0235-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-005-0235-1