Abstract
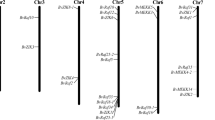
Sulfate transporter (SULTR) proteins are in charge of the transport and absorption on sulfate substances, and have been reported to play vital roles in the biological processes of plant growth and stress response. However, there were few reports of genome-wide identification and expression-pattern analysis of SULTRs in Hibiscus mutabilis. Gossypium genus is a ideal model for studying the allopolyploidy, therefore two diploid species (G. raimondii and G. arboreum) and two tetraploid species (G. hirsutum and G. barbadense) were chosen in this study to perform bioinformatic analyses, identifying 18, 18, 35, and 35 SULTR members, respectively. All the 106 cotton SULTR genes were utilized to construct the phylogenetic tree together with 11 Arabidopsis thaliana, 13 Oryza sativa, and 8 Zea mays ones, which was divided into Group1-Group4. The clustering analyses of gene structures and 10 conserved motifs among the cotton SULTR genes showed the consistent evolutionary relationship with the phylogenetic tree, and the results of gene-duplication identification among the four representative Gossypium species indicated that genome-wide or segment duplication might make main contributions to the expansion of SULTR gene family in cotton. Having conducted the cis-regulatory element analysis in promoter region, we noticed that the existing salicylic acid (SA), jasmonic acid (JA), and abscisic acid (ABA) elements could have influences with expression levels of cotton SULTR genes. The expression patterns of GhSULTR genes were also investigated on the 7 different tissues or organs and the develo** ovules and fibers, most of which were highly expressed in root, stem, sepal, receptacel, ovule at 10 DPA, and fiber at 20 and 25 DPA. In addition, more active regulatory were observed in GhSULTR genes responding to multiple abiotic stresses, and 12 highly expressed genes showed the similar expression patterns in the quantitative Real-time PCR experiments under cold, heat, salt, and drought treatments. These findings broaden our insight into the evolutionary relationships and expression patterns of the SULTR gene family in cotton, and provide the valuable information for further screening the vital candidate genes on trait improvement.








Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Akbudak MA, Filiz E, Kontbay K (2018) Genome-wide identification and cadmium induced expression profiling of sulfate transporter (SULTR) genes in sorghum (Sorghum bicolor L). Biometals 31(1):91–105
Alper SL, Sharma AK (2013) The SLC26 gene family of anion transporters and channels. Mol Aspects Med 34(2–3):494–515
Anjum NA, Gill R, Kaushik M, Hasanuzzaman M, Pereira E, Ahmad I, Tuteja N, Gill SS (2015) ATP-sulfurylase, sulfur-compounds, and plant stress tolerance. Front Plant Sci 6:210
Astolfi S, Celletti S, Vigani G, Mimmo T, Cesco S (2021) Interaction between Sulfur and Iron in plants. Front Plant Sci 12:670308
Aubry E, Dinant S, Vilaine F, Bellini C, Le Hir R (2019) Lateral transport of Organic and Inorganic Solutes. Plants (Basel) 8(1):20
Awazuhara M, Fujiwara T, Hayashi H et al (2010) The function of SULTR2;1 sulfate transporter during seed development in Arabidopsis thaliana. Physiol Plant 125(1):95–105
Buchner P, Parmar S, Kriegel A, Carpentier M, Hawkesford MJ (2010) The sulfate transporter family in wheat: tissue-specific gene expression in relation to nutrition. Mol Plant 3(2):374–389
Cao MJ, Wang Z, Wirtz M, Hell R, Oliver DJ, **ang CB (2013) SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J 73(4):607–616
Cao MJ, Wang Z, Zhao Q, Mao JL, Speiser A, Wirtz M, Hell R, Zhu JK, **ang CB (2014) Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. Plant J 77(4):604–615
Chaudhary S, Sindhu SS, Dhanker R, Kumari A (2023) Microbes-mediated sulphur cycling in soil: impact on soil fertility, crop production and environmental sustainability. Microbiol Res 271:127340
Chen Z, Zhao PX, Miao ZQ, Qi GF, Wang Z, Yuan Y, Ahmad N, Cao MJ, Hell R, Wirtz M, **ang CB (2019) SULTR3s function in Chloroplast Sulfate Uptake and affect ABA biosynthesis and the stress response. Plant Physiol 180(1):593–604
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, **a R (2020) TBtools: an integrative Toolkit developed for interactive analyses of big Biological Data. Mol Plant 13(8):1194–1202
Chen E, Yang X, Liu R, Zhang M, Zhang M, Zhou F, Li D, Hu H, Li C (2022) GhBEE3-Like gene regulated by brassinosteroids is involved in cotton drought tolerance. Front Plant Sci 13:1019146
Dai F, Chen J, Zhang Z, Liu F, Li J, Zhao T, Hu Y, Zhang T, Fang L (2022) COTTONOMICS: a comprehensive cotton multi-omics database. Database (Oxford). :baac080
Du X, Huang G, He S, Yang Z, Sun G, Ma X, Li N, Zhang X, Sun J, Liu M, Jia Y, Pan Z, Gong W, Liu Z, Zhu H, Ma L, Liu F, Yang D, Wang F, Fan W, Gong Q, Peng Z, Wang L, Wang X, Xu S, Shang H, Lu C, Zheng H, Huang S, Lin T, Zhu Y, Li F (2018) Resequencing of 243 diploid cotton accessions based on an updated a genome identifies the genetic basis of key agronomic traits. Nat Genet 50(6):796–802
Dürr J, Bücking H, Mult S, Wildhagen H, Palme K, Rennenberg H, Ditengou F, Herschbach C (2010) Seasonal and cell type specific expression of sulfate transporters in the phloem of Populus reveals tree specific characteristics for SO42– storage and mobilization. Plant Mol Biol 72(4–5):499–517
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39(Web Server issue):W29–37
Gallardo K, Courty PE, Le Signor C, Wipf D, Vernoud V (2014) Sulfate transporters in the plant’s response to drought and salinity: regulation and possible functions. Front Plant Sci 5:580
Ge C, Wang L, Yang Y, Liu R, Liu S, Chen J, Shen Q, Ma H, Li Y, Zhang S, Pang C (2022) Genome-wide association study identifies variants of GhSAD1 conferring cold tolerance in cotton. J Exp Bot 73(7):2222–2237
Hawkesford MJ, De Kok LJ (2006) Managing sulphur metabolism in plants. Plant Cell Environ 29(3):382–395
Heidari P, Hasanzadeh S, Faraji S, Ercisli S, Mora-Poblete F (2023) Genome-wide characterization of the Sulfate Transporter Gene Family in Oilseed crops: Camelina sativa and Brassica napus. Plants (Basel) 12(3):628
Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y, Ju L, Deng J, Zhao T, Lian J, Baruch K, Fang D, Liu X, Ruan YL, Rahman MU, Han J, Wang K, Wang Q, Wu H, Mei G, Zang Y, Han Z, Xu C, Shen W, Yang D, Si Z, Dai F, Zou L, Huang F, Bai Y, Zhang Y, Brodt A, Ben-Hamo H, Zhu X, Zhou B, Guan X, Zhu S, Chen X, Zhang T (2019) Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet 51(4):739–748
Huang Q, Wang M, **a Z (2018) The SULTR gene family in maize (Zea mays L.): gene cloning and expression analyses under sulfate starvation and abiotic stress. J Plant Physiol 220:24–33
Jain M, Tyagi AK, Khurana JP (2006) Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88(3):360–371
Kataoka T, Hayashi N, Yamaya T, Takahashi H (2004a) Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136(4):4198–4204
Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H (2004b) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16(10):2693–2704
Kimura Y, Ushiwatari T, Suyama A, Tominaga-Wada R, Wada T, Maruyama-Nakashita A (2019) Contribution of Root Hair Development to Sulfate Uptake in Arabidopsis. Plants (Basel) 8(4):106
Kirschner S, Woodfield H, Prusko K, Koczor M, Gowik U, Hibberd JM, Westhoff P (2018) Expression of SULTR2;2, encoding a low-affinity sulphur transporter, in the Arabidopsis bundle sheath and vein cells is mediated by a positive regulator. J Exp Bot 69(20):4897–4906
Ku YS, Sintaha M, Cheung MY, Lam HM (2018) Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int J Mol Sci 19(10):3206
Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43(Database issue):D257–D260
Leustek T, Saito K (1999) Sulfate transport and assimilation in plants. Plant Physiol 120(3):637–644
Li PT, Rashid MHO, Chen TT, Lu QW, Ge Q, Gong WK, Liu AY, Gong JW, Shang HH, Deng XY, Li JW, Li SQ, **ao XH, Liu RX, Zhang Q, Duan L, Zou XY, Zhang Z, Jiang X, Zhang Y, Peng RH, Shi YZ, Yuan YL (2019) Transcriptomic and biochemical analysis of upland cotton (Gossypium hirsutum) and a chromosome segment substitution line from G. hirsutum × G. barbadense in response to Verticillium Dahliae infection. BMC Plant Biol 19(1):19
Li Q, Gao Y, Yang A (2020) Sulfur homeostasis in plants. Int J Mol Sci 21(23):8926
Li Z, Liu Z, Wei Y, Liu Y, **ng L, Liu M, Li P, Lu Q, Peng R (2021) Genome-wide identification of the MIOX gene family and their expression profile in cotton development and response to abiotic stress. PLoS ONE 16(7):e0254111
Li Z, Shi Y, **ao X, Song J, Li P, Gong J, Zhang H, Gong W, Liu A, Peng R, Shang H, Ge Q, Li J, Pan J, Chen Q, Lu Q, Yuan Y (2023) Genome-wide characterization of trichome birefringence-like genes provides insights into fiber yield improvement. Front Plant Sci 14:1127760
Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, Lecharny A, Le Ret M, Martin-Magniette ML, Mireau H, Peeters N, Renou JP, Szurek B, Taconnat L, Small I (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16(8):2089–2103
Maharajan T, Krishna TPA, Kiriyanthan RM, Ignacimuthu S, Ceasar SA (2021) Improving abiotic stress tolerance in sorghum: focus on the nutrient transporters and marker-assisted breeding. Planta 254(5):90
Malcheska F, Ahmad A, Batool S, Müller HM, Ludwig-Müller J, Kreuzwieser J, Randewig D, Hänsch R, Mendel RR, Hell R, Wirtz M, Geiger D, Ache P, Hedrich R, Herschbach C, Rennenberg H (2017) Drought-enhanced Xylem Sap Sulfate closes Stomata by affecting ALMT12 and guard cell ABA synthesis. Plant Physiol 174(2):798–814
Malko DB, Makeev VJ, Mironov AA, Gelfand MS (2006) Evolution of exon-intron structure and alternative splicing in fruit flies and malarial mosquito genomes. Genome Res 16(4):505–509
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140(2):411–432
Narayan OP, Verma N, Jogawat A, Dua M, Johri AK (2021) Sulfur transfer from the endophytic fungus Serendipita indica improves maize growth and requires the sulfate transporter SiSulT. Plant Cell 33(4):1268–1285
Narayan OP, Kumar P, Yadav B, Dua M, Johri AK (2023) Sulfur nutrition and its role in plant growth and development. Plant Signal Behav 18(1):2030082
Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP (2002) Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J 29(4):475–486
Song J, Pei W, Wang N, Ma J, **n Y, Yang S, Wang W, Chen Q, Zhang J, Yu J, Wu M, Qu Y (2022) Transcriptome analysis and identification of genes associated with oil accumulation in upland cotton. Physiol Plant 174(3):e13701
Takahashi H (2019) Sulfate transport systems in plants: functional diversity and molecular mechanisms underlying regulatory coordination. J Exp Bot 70(16):4075–4087
Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J 23(2):171–182
Tang Y, Zhang B, Li Z, Deng P, Deng X, Long H, Wang X, Huang K (2023) Overexpression of the sulfate transporter-encoding SULTR2 increases chromium accumulation in Chlamydomonas reinhardtii. Biotechnol Bioeng 120(5):1334–1345
Vatansever R, Koc I, Ozyigit II, Sen U, Uras ME, Anjum NA, Pereira E, Filiz E (2016) Genome-wide identification and expression analysis of sulfate transporter (SULTR) genes in potato (Solanum tuberosum L). Planta 244(6):1167–1183
Waadt R, Seller CA, Hsu PK, Takahashi Y, Munemasa S, Schroeder JI (2022) Plant hormone regulation of abiotic stress responses. Nat Rev Mol Cell Biol 23(10):680–694
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, ** H, Marler B, Guo H, Kissinger JC, Paterson AH (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40(7):e49
Wang W, **e Y, Liu L, King GJ, White P, Ding G, Wang S, Cai H, Wang C, Xu F, Shi L (2022) Genetic control of seed Phytate Accumulation and the development of low-phytate crops: a review and perspective. J Agric Food Chem 70(11):3375–3390
Wen X, Chen Z, Yang Z, Wang M, ** S, Wang G, Zhang L, Wang L, Li J, Saeed S, He S, Wang Z, Wang K, Kong Z, Li F, Zhang X, Chen X, Zhu Y (2023) A comprehensive overview of cotton genomics, biotechnology and molecular biological studies. Sci China Life Sci 66(10):2214–2256
Wilson CM, Yang S, Rodriguez M Jr, Ma Q, Johnson CM, Dice L, Xu Y, Brown SD (2013) Clostridium thermocellum transcriptomic profiles after exposure to furfural or heat stress. Biotechnol Biofuels 6(1):131
Xun M, Song J, Shi J, Li J, Shi Y, Yan J, Zhang W, Yang H (2021) Genome-wide identification of Sultr genes in Malus domestica and Low Sulfur-Induced MhSultr3;1a to increase cysteine-improving growth. Front Plant Sci 12:748242
Yan C, Yang T, Wang B, Yang H, Wang J, Yu Q (2023) Genome-wide identification of the WD40 Gene Family in Tomato (Solanum lycopersicum L). Genes (Basel) 14(6):1273
Yang J, Zhang B, Gu G, Yuan J, Shen S, ** L, Lin Z, Lin J, **e X (2022) Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L). BMC Genomics 23(1):432
Yang L, Wang X, Zhao F, Zhang X, Li W, Huang J, Pei X, Ren X, Liu Y, He K, Zhang F, Ma X, Yang D (2023) Roles of S-Adenosylmethionine and its derivatives in Salt Tolerance of Cotton. Int J Mol Sci 24(11):9517
Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K (2002) Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J 29(4):465–473
Yoshimoto N, Inoue E, Saito K, Yamaya T, Takahashi H (2003) Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol 131(4):1511–1517
Yuan Z, Long W, Hu H, Liang T, Luo X, Hu Z, Zhu R, Wu X (2021) Genome-wide identification and expansion patterns of SULTR Gene Family in Gramineae crops and their expression profiles under abiotic stress in Oryza sativa. Genes (Basel) 12(5):634
Zafar MM, Jia X, Shakeel A, Sarfraz Z, Manan A, Imran A, Mo H, Ali A, Youlu Y, Razzaq A, Iqbal MS, Ren M (2022a) Unraveling Heat Tolerance in Upland Cotton (Gossypium hirsutum L.) using Univariate and Multivariate Analysis. Front Plant Sci 12:727835
Zafar MM, Rehman A, Razzaq A, Parvaiz A, Mustafa G, Sharif F, Mo H, Youlu Y, Shakeel A, Ren M (2022b) Genome-wide characterization and expression analysis of erf gene family in cotton. BMC Plant Biol 22(1):134
Zhang H, Hao X, Zhang J, Wang L, Wang Y, Li N, Guo L, Ren H, Zeng J (2022) Genome-wide identification of SULTR genes in tea plant and analysis of their expression in response to sulfur and selenium. Protoplasma 259(1):127–140
Zhu S, Wang X, Chen W, Yao J, Li Y, Fang S, Lv Y, Li X, Pan J, Liu C, Li Q, Zhang Y (2021) Cotton DMP gene family: characterization, evolution, and expression profiles during development and stress. Int J Biol Macromol 183:1257–1269
Zou J, Hu W, Li Y, Zhu H, He J, Wang Y, Meng Y, Chen B, Zhao W, Wang S, Zhou Z (2022) Leaf anatomical alterations reduce cotton’s mesophyll conductance under dynamic drought stress conditions. Plant J 111(2):391–405
Zuber H, Davidian JC, Wirtz M, Hell R, Belghazi M, Thompson R, Gallardo K (2010) Sultr4;1 mutant seeds of Arabidopsis have an enhanced sulphate content and modified proteome suggesting metabolic adaptations to altered sulphate compartmentalization. BMC Plant Biol 10:78
Funding
This research was funded by the National Natural Science Foundation of China (32272179 and 32070560), the Regional Innovation Guidance Project of **njiang Production and Construction Corps (2021BB012), the Program for Innovative Research Team (in Science and Technology) in University of Henan Province (20IRTSTHN021), the Postgraduate Education Reform and Quality Improvement Project of Henan Province (YJS2022JD47), the National Key R&D Program of China (2021YFE0101200), Zhongyuan Scholars Workstation (224400510020), China Agriculture Research System of MOF and MARA, the National Agricultural Science and Technology Innovation Project for CAAS (CAAS-ASTIP-2016-ICR), Financial science and technology project of Huyanghe City, the seventh Division of **njiang Production and Construction Corps (2012C15), and PhD start-up fund of Anyang Institute of Technology (BSJ2020011).
Author information
Authors and Affiliations
Contributions
Y.C., R.Y., X.X., Y.Y., C.M. and P.L. designed the study; Y.C., R.Y. and X.X. prepared samples and generated the experiments; Y.C. collected and organized data and wrote the manuscript. H.Z., S.Y., B.X., Q.L. Q.L., Y.S., and Y.L. provided suggestions and revised the paper. All authors have read and agreed to the published version of the manuscript. Authorship must be limited to those who have contributed substantially to the work reported.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, Y., **ao, X., Yang, R. et al. Genome-wide identification and expression-pattern analysis of sulfate transporter (SULTR) gene family in cotton under multiple abiotic stresses and fiber development. Funct Integr Genomics 24, 108 (2024). https://doi.org/10.1007/s10142-024-01387-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-024-01387-y