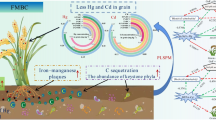
Abstract
Management and improving saline-alkali land is necessary for sustainable agricultural development. We conducted a field experiment to investigate the effects of spraying lactic acid bacteria (LAB) on the cucumber and tomato plant soils. Three treatments were designed, including spraying of water, viable or sterilized LAB preparations to the soils of cucumber and tomato plants every 20 days. Spraying sterilized or viable LAB could reduce the soil pH, with a more obvious effect by using viable LAB, particularly after multiple applications. Metagenomic sequencing revealed that the soil microbiota in LAB-treated groups had higher alpha-diversity and more nitrogen-fixing bacteria compared with the water-treated groups. Both viable and sterilized LAB, but not water application, increased the complexity of the soil microbiota interactive network. The LAB-treated subgroups were enriched in some KEGG pathways compared with water or sterilized LAB subgroups, such as environmental information processing–related pathways in cucumber plant; and metabolism-related pathways in tomato plant, respectively. Redundancy analysis revealed association between some soil physico-chemical parameters (namely soil pH and total nitrogen) and bacterial biomarkers (namely Rhodocyclaceae, Pseudomonadaceae, Gemmatimonadaceae, and Nitrosomonadales). Our study demonstrated that LAB is a suitable strategy for decreasing soil pH and improving the microbial communities in saline-alkali land.






Similar content being viewed by others
Data availability
The entire sequence data set has been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (Accession No: SRR16229770-SRR16229787). All data analyzed during this study are included in the additional files of this published article.
Abbreviations
- LAB :
-
Lactic acid bacteria
- PCoA :
-
Principal coordinates analysis
- NMDS :
-
Nonmetric multidimensional scales ordination
- RDA :
-
Redundancy analysis
- PICRUSt :
-
Phylogenetic investigation of communities by reconstruction of unobserved states
- PPM :
-
Plant probiotic microorganisms
- OTUs :
-
Operational taxonomic units
- LEfSe :
-
Linear discriminant analysis (LDA) effect size
- KEGG :
-
Kyoto Encyclopedia of Genes and Genomes
- KO :
-
KEGG Ontology
References
Ahmed V, Verma MK, Gupta S et al (2018) Metagenomic profiling of soil microbes to mine salt stress tolerance genes. Front Microbiol [J] 9:159. https://doi.org/10.3389/fmicb.2018.00159
Aslam R, Bostan N, Nabgha-e-Amen et al (2011) A critical review on halophytes: salt tolerant plants. J Med Plants Res [J] 5:7108–7118. https://doi.org/10.5897/jmprx11.009
Aulakh AM, Qadir G, Hassan FU et al (2020) Desert soil microbes as a mineral nutrient acquisition tool for chickpea (Cicer arietinum L.) productivity at different moisture regimes. Plants (Basel) [J] 9:1629–1650. https://doi.org/10.3390/plants9121629
Bachran M, Kluge S, Lopez-Fernandez M et al (2019) Microbial diversity in an arid, naturally saline environment. Microb Ecol [J] 78:494–505. https://doi.org/10.1007/s00248-018-1301-2
Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks [M], Third international AAAI conference on weblogs and social media.https://doi.org/10.13140/2.1.1341.1520
Bhardwaj D, Ansari MW, Sahoo RK et al (2014) Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb Cell Factories [J] 13:66–76
Brockett BFT, Prescott CE, Grayston SJ (2012) Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol Biochem [J] 44:9–20. https://doi.org/10.1016/j.soilbio.2011.09.003
Canfora L, Bacci G, Pinzari F et al (2014) Salinity and bacterial diversity: to what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS One [J] 9:e106662. https://doi.org/10.1371/journal.pone.0106662
Chen YS, Yanagida F, Shinohara T (2005) Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett Appl Microbiol [J] 40:195–200. https://doi.org/10.1111/j.1472-765X.2005.01653.x
Chen YS, Wu HC, Lo HY et al (2012) Isolation and characterisation of lactic acid bacteria from jiang-gua (fermented cucumbers), a traditional fermented food in Taiwan. J Sci Food Agric [J] 92:2069–2075. https://doi.org/10.1002/jsfa.5583
Chen Q-L, Ding J, Zhu D et al (2020) Rare microbial taxa as the major drivers of ecosystem multifunctionality in long-term fertilized soils. Soil Biol Biochem [J] 141:107686. https://doi.org/10.1016/j.soilbio.2019.107686
Daniel R (2005) The metagenomics of soil. Nat Rev Microbiol [J] 3:470–478. https://doi.org/10.1038/nrmicro1160
de Souza Vandenberghede LP, Blandon Garcia LM, Rodrigues C et al (2017) Potential applications of plant probiotic microorganisms in agriculture and forestry. AIMS Microbiol [J] 3:629–648. https://doi.org/10.3934/microbiol.2017.3.629
Delgado-Baquerizo M, Oliverio AM, Brewer TE et al (2018) A global atlas of the dominant bacteria found in soil. Science [J] 359:320. https://doi.org/10.1126/science.aap9516
Dutta T, Rahman MM, Bhuiya MSU et al (2015) Use of organic amendment for amelioration of salinity stress in transplanted aman rice cv. BRRI dhan41. Int J Nat Soc Sci [J] 2(5):82–94
Fierer N, Leff JW, Adams BJ et al (2012) Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A [J] 109:21390–21395. https://doi.org/10.1073/pnas.1215210110
Garbeva P, van Veen JA, van Elsas JD (2004) Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol [J] 42:243–270. https://doi.org/10.1146/annurev.phyto.42.012604.135455
Garg N, Saroy K, Cheema A et al (2019) Microbial diversity in soil: biological tools for abiotic stress management in plants[C]//plant biotic interactions.Springer Nature Switzerland AG. https://doi.org/10.1007/978-3-030-26657-8_17
Glick BR (1995) The enhancement of plant growth by free living bacteria. Can J Microbiol [J] 41:109–117. https://doi.org/10.1139/m95-015
Glick BR, Penrose DM, Li J (1998) A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J Theor Biol [J] 190:63–68. https://doi.org/10.1006/jtbi.1997.0532
Hayat R, Ali S, Amara U et al (2010) Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol [J] 60:579–598. https://doi.org/10.1007/s13213-010-0117-1
Hou J, Wang X, Chen Y et al (2020) Study on the effect of lactic acid bacteria compound fertilizer in saline-alkali soil fertilization and promoting vegetable growth. J Inner Mongolia Agric Univ (Nat Sci Ed) [J] 41:37–43
Jia Y, Whalen JK (2020) A new perspective on functional redundancy and phylogenetic niche conservatism in soil microbial communities. Pedosphere [J] 30:18–24. https://doi.org/10.1016/s1002-0160(19)60826-x
**g G, Sun Z, Wang H et al (2017) Parallel-META 3: comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities. Sci Rep [J] 7. https://doi.org/10.1038/srep40371
Jousset A, Bienhold C, Chatzinotas A et al (2017) Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J [J] 11:853–862. https://doi.org/10.1038/ismej.2016.174
Kang S-M, Radhakrishnan R, You Y-H et al (2014) Cucumber performance is improved by inoculation with plant growth-promoting microorganisms. Acta Agric Scand Sect B Soil Plant Sci [J] 65:36–44. https://doi.org/10.1080/09064710.2014.960889
Ladygina N, Hedlund K (2010) Plant species influence microbial diversity and carbon allocation in the rhizosphere. Soil Biol Biochem [J] 42:162–168. https://doi.org/10.1016/j.soilbio.2009.10.009
Lamont JR, Wilkins O, Bywater-Ekegärd M et al (2017) From yogurt to yield: potential applications of lactic acid bacteria in plant production. Soil Biol Biochem [J] 111:1–9. https://doi.org/10.1016/j.soilbio.2017.03.015
Lauber CL, Hamady M, Knight R et al (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol [J] 75:5111–5120. https://doi.org/10.1128/AEM.00335-09
Li K, Zhang W, Kwok L-Y et al (2020) Screening of Lactobacillus plantarum with broad-spectrum antifungal activity and its application in preservation of golden-red apples. Czech J Food Sci [J] 38:315–322. https://doi.org/10.17221/175/2020-cjfs
Li L, Li J, Zhu S (2022) Research status of different improvement measures for saline-alkali land. E3S Web Conf [J]. 338. https://doi.org/10.1051/e3sconf/202233801038
Louca S, Polz MF, Mazel F et al (2018) Function and functional redundancy in microbial systems. Nat Ecol Evol [J] 2:936–943. https://doi.org/10.1038/s41559-018-0519-1
Mahanty T, Bhattacharjee S, Goswami M et al (2017) Biofertilizers: a potential approach for sustainable agriculture development. Environ Sci Pollut Res [J] 24:3315–3335. https://doi.org/10.1007/s11356-016-8104-0
Makarovaa K, Slesarevb A, Wolfa Y et al (2006) Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A [J] 103(42):15611–15616
Mccune B, Grace JB (2002) Analysis of ecological communities. MJM Software Design, Gleneden Beach
Miyajima S, Uoi N, Murata T et al (2015) Effect of structural modification on heat transfer through man-made soils in urban green areas. Soil Sci Plant Nutr [J] 61:70–87. https://doi.org/10.1080/00380768.2015.1051929
Mocali S, Benedetti A (2010) Exploring research frontiers in microbiology: the challenge of metagenomics in soil microbiology. Res Microbiol [J] 161:497–505. https://doi.org/10.1016/j.resmic.2010.04.010
Numan M, Bashir S, Khan Y et al (2018) Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review. Microbiol Res [J] 209:21–32. https://doi.org/10.1016/j.micres.2018.02.003
Olasupo NA (1996) Bacteriocins of Lactobacillus plantarum strains from fermented foods. Folia Microbiol [J] 41(2):130–136. https://doi.org/10.1007/BF02814687
Oren A (2014) The family Rhodocyclaceae[C]//the prokaryotes. 975–998. https://doi.org/10.1007/978-3-642-30197-1_292
Pankhurst CE, Yu S, Hawke BG et al (2001) Capacity of fatty acid profiles and substrate utilization patterns to describe differences in soil microbial communities associated with increased salinity or alkalinity at three locations in South Australia. Biol Fertil Soils [J] 33(3):204–217
Qiu L, Zhang Q, Zhu H et al (2021) Erosion reduces soil microbial diversity, network complexity and multifunctionality. ISME J [J] 15:2474–2489. https://doi.org/10.1038/s41396-021-00913-1
Rousk J, Baath E, Brookes PC et al (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J [J] 4:1340–1351. https://doi.org/10.1038/ismej.2010.58
Segata N, Izard J, Walron L et al (2011) Metagenomic biomarker discovery and explanation. Genome Biol [J] 12(6):R60. https://doi.org/10.1186/gb-2011-12-6-r60
Shao T, Gu X, Zhu T et al (2019) Industrial crop Jerusalem artichoke restored coastal saline soil quality by reducing salt and increasing diversity of bacterial community. Appl Soil Ecol [J] 138:195–206. https://doi.org/10.1016/j.apsoil.2019.03.003
Shrivastava P, Kumar R (2015) Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci [J] 22:123–131. https://doi.org/10.1016/j.sjbs.2014.12.001
Su X, Xu J, Ning K (2012) Parallel-META: efficient metagenomic data analysis based on high-performance computation. BMC Syst Biol [J] 6(Suppl 1):S16. https://doi.org/10.1186/1752-0509-6-S1-S16
Su X, Pan W, Song B et al (2014) Parallel-META 2.0: enhanced metagenomic data analysis with functional annotation, high performance computing and advanced visualization. PLoS One [J] 9:e89323. https://doi.org/10.1371/journal.pone.0089323
van der Heijden Marcel GA, Bardgett Richard D, van Straalen Nico M (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett [J] 11:296–310. https://doi.org/10.1111/j.1461-0248.2007.01139.x
Vargas R, Krasilnikov P, Balyuk SA et al (2018) Handbook for saline soil management. Eurasian Soil Partnership implementation plan[C]//:Food and Agriculture Organization of the United Nations and Lomonosov Moscow State University
Wang H, Yan H, Shin J et al (2011) Activity against plant pathogenic fungi of Lactobacillus plantarum IMAU10014 isolated from **njiang koumiss in China. Ann Microbiol [J] 61:879–885. https://doi.org/10.1007/s13213-011-0209-6
Wang S, Feng Q, Zhou Y et al (2017) Dynamic changes in water and salinity in saline-alkali soils after simulated irrigation and leaching. PLoS One [J] 12:e0187536. https://doi.org/10.1371/journal.pone.0187536
Wang Q, Cao Z, Liu Q et al (2019) Enhancement of COD removal in constructed wetlands treating saline wastewater: intertidal wetland sediment as a novel inoculation. J Environ Manag [J] 249:109398. https://doi.org/10.1016/j.jenvman.2019.109398
Wang J, Shi X, Zheng C et al (2021) Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci Total Environ [J] 755:142449. https://doi.org/10.1016/j.scitotenv.2020.142449
Wolińska A (2019) Metagenomic achievements in microbial diversity determination in croplands[C]//microbial diversity in the genomic era. 15–35. https://doi.org/10.1016/b978-0-12-814849-5.00002-2
Xun W, Huang T, Zhao J et al (2015) Environmental conditions rather than microbial inoculum composition determine the bacterial composition, microbial biomass and enzymatic activity of reconstructed soil microbial communities. Soil Biol Biochem [J] 90:10–18. https://doi.org/10.1016/j.soilbio.2015.07.018
Yin J, Liu W (2019) Status and treatment of saline-alkali soils in Tumd Left Banner of Inner Mongolia Autonomous Region. Ningxia J Agric For Sci Tech [J] 60:40–41. https://doi.org/10.3969/j.issn.1002-204x.2019.11.014
Zeng Q, Dong Y, An S (2016) Bacterial community responses to soils along a latitudinal and vegetation gradient on the Loess Plateau, China. PLoS One [J] 11:e0152894. https://doi.org/10.1371/journal.pone.0152894
Zhang JG, Cai Y, Kobayashi R et al (2000) Characteristics of lactic acid bacteria isolated from forage crops and their effects on silage fermentation. J Sci Food Agric [J] 80:1455–1460. https://doi.org/10.1002/1097-0010(200008)80:103.0.CO;2-C
Zhang J, Liu W, Sun Z et al (2011) Diversity of lactic acid bacteria and yeasts in traditional sourdoughs collected from western region in Inner Mongolia of China. Food Control [J] 22:767–774. https://doi.org/10.1016/j.foodcont.2010.11.012
Zhao L, Heng T, Yang L et al (2021) Study on the farmland improvement effect of drainage measures under film mulch with drip irrigation in saline–alkali land in arid areas. Sustainability [J] 13:4159. https://doi.org/10.3390/su13084159
Zhu JK (2001) Plant salt tolerence. Trands Plant Sci [J] 6:66–71. https://doi.org/10.1016/S1360-1385(00)01838-0
Zlotnikov KM, Zlotnikovb AK, Kaparullinaa EN et al (2013) Phylogenetic position and phosphate solubilization activity of lactic acid bacteria associated with different plants. Microbiology [J] 82:393–396. https://doi.org/10.1134/S0026261713030144
Funding
This study was supported by the Science and Technology Project of College of Food Science and Engineering, Inner Mongolia Agricultural University (SPKJ201905).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: ZZ; MB.
Performed field experiments and managed the project: JQH; MB.
Analyzed data: HT, LJY, ZXZ.
Wrote the manuscript: HT.
Critical evaluation and extensive revision of the manuscript: LYK.
All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tang, H., Zhong, Z., Hou, J. et al. Metagenomic analysis revealed the potential of lactic acid bacteria in improving natural saline-alkali land. Int Microbiol 27, 311–324 (2024). https://doi.org/10.1007/s10123-023-00388-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-023-00388-4