Abstract
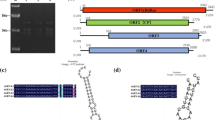
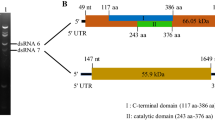
Beauveria bassiana Vuillemin is an entomopathogenic fungus that has been developed as a biological insecticide. B. bassiana can be infected by single or multiple mycoviruses, most of which are double-stranded RNA (dsRNA) viruses, while infections with single-stranded RNA (ssRNA) viruses, especially negative single-stranded RNA (-ssRNA) viruses, have been observed less frequently. In the present study, we sequenced and analyzed the complete genomes of two new different mycoviruses coinfecting a single B. bassiana strain: a -ssRNA virus which we have named "Beauveria bassiana negative-strand RNA virus 1" (BbNSRV1), and a dsRNA virus, which we have named "Beauveria bassiana orthocurvulavirus 1" (BbOCuV1). The genome of BbNSRV1 consists of a single segment of negative-sense, single-stranded RNA with a length of 6169 nt, containing a single open reading frame (ORF) encoding a putative RNA-dependent RNA polymerase (RdRp) with 1949 aa (220.1 kDa). BLASTx analysis showed that the RdRp had the highest sequence similarity (59.79%) to that of Plasmopara viticola lesion associated mononegaambi virus 2, a member of the family Mymonaviridae. This is the first report of a -ssRNA mycovirus infecting B. bassiana. The genome of BbOCuV1 consists of two dsRNA segments, 2164 bp and 1765 bp in length, respectively, with dsRNA1 encoding a protein with conserved RdRp motifs and 70.75% sequence identity to the putative RdRp of the taxonomically unassigned mycovirus Fusarium graminearum virus 5 (FgV5), and the dsRNA2 encoding a putative coat protein with sequence identity 64.26% to the corresponding protein of the FgV5. Phylogenetic analysis indicated that BbOCuV1 belongs to a taxonomically unassigned group of dsRNA mycoviruses related to members of the families Curvulaviridae and Partitiviridae. Hence, it might be the member of a new family that remains to be named and formally recognized.


Similar content being viewed by others
References
Kotta-Loizou I, Coutts RHA (2017) Studies on the virome of the entomopathogenic fungus Beauveria bassiana reveal novel dsRNA elements and mild hypervirulence. PLOS Pathog 13:e1006183. https://doi.org/10.1371/journal.ppat.1006183
Li L, Kang Q, Zhang SB et al (2021) The whole genome sequence of a novel chrysovirus from Beauveria Bassiana Vuillemin, an entomopathogenic fungus. Arch Virol 3443–3447. https://doi.org/10.21203/rs.3.rs-488047/v1
Kang Q, Li L, Li J et al (2021) A novel polymycovirus with defective RNA isolated from the entomopathogenic fungus Beauveria bassiana Vuillemin. Arch Virol 166:3487–3492. https://doi.org/10.1007/s00705-021-05238-0
Filippou C, Garrido-Jurado I, Meyling NV et al (2018) Mycoviral population dynamics in Spanish isolates of the entomopathogenic fungus Beauveria bassiana. Viruses 10:665. https://doi.org/10.3390/v10120665
Shi N, Yang G, Wang P et al (2019) Complete genome sequence of a novel partitivirus from the entomogenous fungus Beauveria bassiana in China. Arch Virol 164:3141–3144. https://doi.org/10.1007/s00705-019-04428-1
Shi N, Hu F, Wang P et al (2021) Molecular characterization of two dsRNAs that could correspond to the genome of a new mycovirus that infects the entomopathogenic fungus Beauveria bassiana. Arch Virol 166:3233–3237. https://doi.org/10.1007/s00705-021-05239-z
Koloniuk I, Hrabáková L, Petrzik K (2015) Molecular characterization of a novel amalgavirus from the entomopathogenic fungus Beauveria bassiana. Arch Virol 160:1585–1588. https://doi.org/10.1007/s00705-015-2416-0
Kondo H, Chiba S, Toyoda K, Suzuki N et al (2013) Evidence for negative-strand RNA virus infection in fungi. Virology 435:201–209. https://doi.org/10.1016/j.virol.2012.10.002
Liu L, **e J, Cheng J et al (2014) Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci U S A 111:12205–12210. https://doi.org/10.1073/pnas.1401786111
Jiang et al (2022) ICTV Virus Taxonomy Profile: Mymonaviridae 2022. JGV 103. https://doi.org/10.1099/jgv.0.001787
Chiapello M, Rodríguez-Romero J, Ayllón MA et al (2020) Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol 6:veaa058. https://doi.org/10.1093/ve/veaa058
Morris TJ, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858. https://doi.org/10.1094/Phyto-69-854
Pankovics P, Boros Á, Reuter G (2015) Novel 5’/3'RACE method for amplification and determination of single-stranded RNAs through double-stranded RNA (dsRNA) intermediates. Mol Biotechnol 57:974–981. https://doi.org/10.1007/s12033-015-9889-7
Darissa O, Willingmann P, Adam G (2010) Optimized approaches for the sequence determination of double-stranded RNA templates. J Virol Methods 169:397–403. https://doi.org/10.1016/j.jviromet.2010.08.013
Cho WK, Lee KM, Yu J et al (2013) Insight into mycoviruses infecting Fusarium species. Adv Virus Res 86:273–288. https://doi.org/10.1016/b978-0-12-394315-6.00010-6
Wang L et al (2017) The complete genome sequence of a double-stranded RNA mycovirus from Fusarium Graminearum Strain Hn1. Arch Virol 7:2119–2124. https://doi.org/10.1007/s00705-017-3317-1
Acknowledgments
This study was supported by Jilin Project for Outstanding Talents (Teams) for Innovation and Entrepreneurship in Science and Technology (grant number 20230508011RC).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical approval
This article does not contain any studies involving human participants or animals.
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Communicated by Stephen John Wylie
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, M., Liu, H., Jia, X. et al. The complete genome sequences of a negative single-stranded RNA virus and a double-stranded RNA virus coinfecting the entomopathogenic fungus Beauveria bassiana Vuillemin. Arch Virol 169, 42 (2024). https://doi.org/10.1007/s00705-024-05985-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-024-05985-w