Abstract
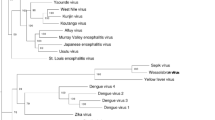
Menghai flavivirus (MFV) was isolated from Aedes albopictus in Menghai county of Yunnan Province, China, during an arboviruses screening program in August 2010. Whole genome sequencing of MFV was performed using an Ion PGM™ Sequencer. The complete genome of MFV was 10897 nucleotides in length and encoded a polyprotein and fairly interesting flavivirus orf (FIFO). The polyprotein contained three flavivirus structural proteins (C, prM/M and E) and seven nonstructural proteins. Nucleotide BLAST analysis revealed that the MFV genome showed highest similarity to **shuangbanna Aedes flavivirus, a novel insect-specific flavivirus recently isolated from the same area. These species shared a query cover of 99%, but only 71% identity, while FIFO showed no similarity with any of the published sequences. Genomic and phylogenetic analyses suggested that MFV was a novel species of the genus Flavivirus. Our findings enrich our understanding of the genetics and prevalence of the family Flaviviridae.

Similar content being viewed by others
References
Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P (2016) Genetic dissection of flaviviridae host factors through genome-scale CRISPR screens. Nature. doi:10.1038/nature18631
Wang HJ, Li XF, Liu L, Xu YP, Ye Q, Deng YQ, Huang XY, Zhao H, Qin ED, Shi PY (2016) The emerging duck flavivirus is not pathogenic for primates and is highly sensitive to mammal interferon signaling. J Virol. doi:10.1128/JVI.00197-16
Crabtree MB, Nga PT, Miller BR (2009) Isolation and characterization of a new mosquito flavivirus, Quang Binh virus, from Vietnam. Arch Virol 154:857–860
Mclean BJ, Hobson-Peters J, Webb CE, Watterson D, Prow NA, Hong DN, Hall-Mendelin S, Warrilow D, Johansen CA, Jansen CC (2015) A novel insect-specific flavivirus replicates only in aedes -derived cells and persists at high prevalence in wild Aedes vigilax populations in Sydney, Australia. Virology 486:272–283
Hang F, Zhao Q, Guo X, Qiang S, Zuo S, Chao W, Zhou H, An X, Pei G, Tong Y (2016) Complete genome sequence of xishuangbanna flavivirus, a novel mosquito-specific flavivirus from china. Arch Virol 161(6):1723–1727. doi:10.1007/s00705-016-2827-6
Maher-Sturgess SL, Forrester NL, Wayper PJ, Gould EA, Hall RA, Barnard RT, Gibbs MJ (2008) Universal primers that amplify RNA from all three flavivirus subgroups. Virol J 5:1–10
Patel RK, Jain M (2012) Ngs qc toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619
Li S, Fan H, An X, Fan H, Jiang H, Chen Y, Tong Y (2014) Scrutinizing virus genome termini by high-throughput sequencing. PLoS One 9:e85806
Zhang X, Wang Y, Li S, An X, Pei G, Huang Y, Fan H, Mi Z, Zhang Z, Wang W et al (2015) A novel termini analysis theory using hts data alone for the identification of enterococcus phage ef4-like genome termini. BMC Genomics 16:414
Odelius K, Finne A, Albertsson AC (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432
Tang B, Wang Q, Yang M, **e F, Zhu Y, Zhuo Y, Wang S, Gao H, Ding X, Zhang L (2013) Contigscape: a cytoscape plugin facilitating microbial genome gap closing. BMC Genomics 14:1–12
Aziz RK, Bartels D, Best AA, Dejongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB (1998) Phylogeny of the genus flavivirus. J Virol 72:73–83
Firth AE, Blitvich BJ, Wills NM, Miller CL, Atkins JF (2010) Evidence for ribosomal frameshifting and a novel overlap** gene in the genomes of insect-specific flaviviruses. Virology 399:153–166
Blitvich BJ, Firth AE (2015) Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses 7:1927–1959
Fan H, Zhao Q, Guo X, Sun Q, Zuo S, Wu C, Zhou H, An X, Pei G, Tong Y et al (2016) Complete genome sequence of xishuangbanna flavivirus, a novel mosquito-specific flavivirus from china. Arch Virol 161:1723–1727
Patkar CG, Jones CT, Chang YH, Warrier R, Kuhn RJ (2007) Functional requirements of the yellow fever virus capsid protein. J Virol 81:6471–6481
Lin YJ, Wu SC (2005) Histidine at residue 99 and the transmembrane region of the precursor membrane prm protein are important for the prm-e heterodimeric complex formation of japanese encephalitis virus. J Virol 79:8535–8544
Murray CL, Jones CT, Rice CM (2008) Architects of assembly: roles of flaviviridae non-structural proteins in virion morphogenesis. Nat Rev Microbiol 6:699–708
Leung JY, Pijlman GP, Kondratieva N, Hyde J, Mackenzie JM, Khromykh AA (2008) Role of nonstructural protein ns2a in flavivirus assembly. J Virol 82:4731–4741
Lindenbach BD, Rice CM (1999) Genetic interaction of flavivirus nonstructural proteins ns1 and ns4a as a determinant of replicase function. J Virol 73:4611–4621
Liu WJ, Chen HB, Wang XJ, Huang H, Khromykh AA (2004) Analysis of adaptive mutations in kun** virus replicon rna reveals a novel role for the flavivirus nonstructural protein ns2a in inhibition of beta interferon promoter-driven transcription. J Virol 78:12225–12235
Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM (1993) Characterization of the hepatitis c virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol 67:2832–2843
Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Sui SY, Wang CH, Lim SP, Strongin A (2008) Insights into RNA unwinding and atp hydrolysis by the flavivirus ns3 protein. EMBO J 27:3209–3219
Wicker JA, Whiteman MC, Beasley DW, Davis CT, McGee CE, Lee JC, Higgs S, Kinney RM, Huang CY, Barrett AD (2012) Mutational analysis of the west Nile virus ns4b protein. Virology 426:22–33
Kamer G, Argos P (1984) Primary structural comparison of rna-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res 12:7269–7282
Miller RH, Purcell RH (1990) Hepatitis c virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA 87:2057–2061
Katoh K, Standley DM (2013) Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This research was supported by grants from the National Natural Science Foundation of China (No. 81273138 and No. 81572045), the China Mega-Project on Infectious Disease Prevention (No. 2013ZX10004-605, No. 2013ZX10004-607, No. 2013ZX10004-217 and No. 2011ZX10004-001) and the National Hi-Tech Research and Development (863) Program of China (No. 2014AA021402, No. 2012AA022-003 and No. 2015AA020108).
Conflict of interest
All authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals.
Additional information
X. Zhang and X. Guo contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, X., Guo, X., Fan, H. et al. Complete genome sequence of Menghai flavivirus, a novel insect-specific flavivirus from China. Arch Virol 162, 1435–1439 (2017). https://doi.org/10.1007/s00705-017-3232-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3232-5