Abstract
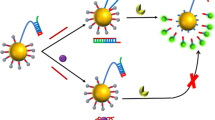
A precisely designed dual-color biosensor has realized a visual assessment of thymidine kinase 1 (TK1) mRNA in both living cells and cell lysates. The oligonucleotide probe is constructed by hybridizing the antisense strand of the target and two recognition sequences, in which FAM serves as the donor and TAMRA as the acceptor. Once interacting with the target, two recognition strands are replaced, and then the antisense complementary sequence forms a more stable double-stranded structure. Due to the increasing spatial distance between two dyes, the FRET is attenuated, leading to a rapid recovery of FAM fluorescence and a reduction of TAMRA fluorescence. A discernible color response from orange to green could be observed by the naked eye, with a limit of detection (LOD) of 0.38 nM and 5.22 nM for spectrometer- and smartphone-based assays, respectively. The proposed ratiometric method transcends previous reports in its capacities in visualizing TK1 expression toward reliable nucleic acid biomarker analysis, which might establish a general strategy for ratiometric biosensing via strand displacement.
Graphical abstract








Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Aufderklamm S, Todenhöfer T, Gakis G et al (2012) Thymidine kinase and cancer monitoring. Cancer Lett 316(1):6–10. https://doi.org/10.1016/j.canlet.2011.10.025
Sawyers CL (2008) The cancer biomarker problem. Nature 452:548–552. https://doi.org/10.1038/nature06913
Nikanjam M, Kato S, Kurzrock R (2022) Liquid biopsy: current technology and clinical applications. J Hematol Oncol 15:131. https://doi.org/10.1186/s13045-022-01351-y
McCartney A, Malorni L (2020) Thymidine kinase-1 as a biomarker in breast cancer: estimating prognosis and early recognition of treatment resistance. Biomark Med 14:495–498. https://doi.org/10.2217/bmm-2020-0072
Jagarlamudi KK, Shaw M (2018) Thymidine kinase 1 as a tumor biomarker: technical advances offer new potential to an old biomarker. Biomark Med 12:1035–1048. https://doi.org/10.2217/bmm-2018-0157
Bitter EE, Townsend MH, Erickson R et al (2020) Thymidine kinase 1 through the ages: a comprehensive review. Cell Biosci 10:138. https://doi.org/10.1186/s13578-020-00493-1
Roy R, Hohng S, Ha T (2008) A practical guide to single-molecule FRET. Nat Methods 5:507–516. https://doi.org/10.1038/nmeth.1208
Larkey NE, Zhang LL, Lansing SS et al (2016) Forster resonance energy transfer to impart signal-on and -off capabilities in a single microRNA biosensor. Analyst 141:6239–6250. https://doi.org/10.1039/D0AY01961E
Yang YJ, Huang J, Yang XH et al (2015) FRET nanoflares for intracellular mRNA detection: avoiding false positive signals and minimizing effects of System fluctuations. J Am Chem Soc 137:8340–8343. https://doi.org/10.1021/jacs.5b04007
Tsourkas A, Behlke MA, Xu Y, Bao G (2003) Spectroscopic features of dual fluorescence/luminescence resonance energy-transfer molecular beacons. Anal Chem 75:3697–3703. https://doi.org/10.1021/ac034295l
Karbownik M, Brzezianska E, Lewinski A (2005) Increased expression of mRNA specific for thymidine kinase, deoxycytidine kinase or thymidine phosphorylase in human papillary thyroid carcinoma. Cancer Lett 225:267–273. https://doi.org/10.1016/j.canlet.2004.10.044
Koczor CA, Torres RA, Fields EJ et al (2013) Thymidine kinase and mtDNA depletion in human cardiomyopathy: epigenetic and translational evidence for energy starvation. Physiol Genomics 45:590–596. https://doi.org/10.1152/physiolgenomics.00014.2013
Jeong MH, ** YH, Kang EY et al (2004) The modulation of radiation-induced cell death by genistein in K562 cells: activation of thymidine kinase 1. Cell Res 14:295–302. https://doi.org/10.1038/sj.cr.7290230
Sun Z, Tong Y, Zhou X et al (2021) Ratiometric fluorescent biosensor based on Forster resonance energy transfer between carbon dots and acridine Orange for miRNA analysis. ACS Omega 6:34150–34159. https://doi.org/10.1021/acsomega.1c05901
Fu Y, Chen T, Wang G et al (2017) Production of a fluorescence resonance energy transfer (FRET) biosensor membrane for microRNA detection. J Mater Chem B 5:7133–7139. https://doi.org/10.1039/C7TB01399J
Chen J, Zhang Y, Chen D et al (2021) Toehold-mediated ligation-free rolling circle amplification enables sensitive and rapid imaging of messenger RNAs in situ in cells. Anal Chim Acta 1160:338463. https://doi.org/10.1016/j.aca.2021.338463
Li T, Sun M, **a S et al (2024) A binary system based DNA tetrahedron and fluorogenic RNA aptamers for highly specific and label-free mRNA imaging in living cells. Talanta 269:125465. https://doi.org/10.1016/j.talanta.2023.125465
Huang J, Wang H, Yang X et al (2016) Fluorescence resonance energy transfer-based hybridization chain reaction for in situ visualization of tumor-related mRNA. Chem Sci 7:3829–3835. https://doi.org/10.1039/C6SC00377J
Mo L, Liang D, He W et al (2021) Ratiometric and amplified fluorescence nanosensor based on a DNA tetrahedron for miRNA imaging in living cells. J Mater Chem B 9:8341–8347. https://doi.org/10.1039/D1TB01537K
Zhao T, Yan W, Dong F et al (2022) A smartphone-based platform for ratiometric visualization of SARS-CoV-2 via an oligonucleotide probe. Microchim Acta 189:268. https://doi.org/10.1007/s00604-022-05364-9
Qian L, Winfree E, Bruck J (2011) Neural network computation with DNA strand displacement cascades. Nature 475:368–372. https://doi.org/10.1038/nature10262
Rogers WB, Manoharan VN (2015) Programming colloidal phase transitions with DNA strand displacement. Science 347:639–642. https://doi.org/10.1126/science.1259762
Bielec K, Sozanski K, Seynen M et al (2019) Kinetics and equilibrium constants of oligonucleotides at low concentrations. Hybridization and melting study. Phys Chem Chem Phys 21:10798–10807. https://doi.org/10.1039/C9CP01295H
Zhang Y, Liang Z, Zong Y et al (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7:12617. https://doi.org/10.1038/ncomms12617
Juma KM, Takita T, Ito K et al (2021) Optimization of reaction condition of recombinase polymerase amplification to detect SARS-CoV-2 DNA and RNA using a statistical method. Biochem Bioph Res Co 567:195–200. https://doi.org/10.1016/j.bbrc.2021.06.023
Ma XP, Wang YG, Zhao T et al (2014) Ultra-pH-sensitive nanoprobe Library with broad pH tunability and fluorescence emissions. J Am Chem Soc 136:11085–11092. https://doi.org/10.1021/ja5053158
Alaejos ÁC, Cabrera SJ, Rodríguez BC, Castañeda AZ (2021) Validation and comparison of two analytical methods for imatinib therapeutic drug monitoring. Chromatographia 84:589–596. https://doi.org/10.1007/s10337-021-04041-y
Ou M, Huang J, Yang XH et al (2017) MnO2 nanosheet mediated DD-A FRET binary probes for sensitive detection of intracellular mRNA. Chem Sci 8:668–673. https://doi.org/10.1039/C6SC03162E
Ratajczak K, Krazinski BE, Kowalczyk AE et al (2018) Hairpin-hairpin molecular beacon interactions for detection of survivin mRNA in malignant SW480 cells. ACS Appl Mater Inter 10:17028–17039. https://doi.org/10.1021/acsami.8b02342
Jiang HY, Li FR, Li W et al (2018) Multiplexed determination of intracellular messenger RNA by using a graphene oxide nanoprobe modified with target-recognizing fluorescent oligonucleotides. Microchim Acta 185:552. https://doi.org/10.1007/s00604-018-3090-1
Fan JL, Tong CY, Dang WY et al (2019) An rGONS-based biosensor for simultaneous imaging of p53 and p21 mRNA in living cells. Talanta 204:20–28. https://doi.org/10.1016/j.talanta.2019.05.087
Zhang WY, Hao WH, Liu XT et al (2020) Visual detection of miRNAs using enzyme-free amplification reactions and ratiometric fluorescent probes. Talanta 219:121332. https://doi.org/10.1016/j.talanta.2020.121332
Borghei YS, Hosseini M, Ganjali MR, Ju HX (2018) Colorimetric and energy transfer based fluorometric turn-on method for determination of microRNA using silver nanoclusters and gold nanoparticles. Microchim Acta 185:286. https://doi.org/10.1007/s00604-018-2825-3
Yang JC, Wang K, Xu H et al (2019) Detection platforms for point-of-care testing based on colorimetric, luminescent and magnetic assays: a review. Talanta 202:96–110. https://doi.org/10.1016/j.talanta.2019.04.054
Yan W, Wang W, Zheng G et al (2023) Two birds with one stone: ratiometric sensing platform overcoming cross-interference for multiple-scenario detection and accurate discrimination of tetracycline analogs. J Hazard Mater 458:132016. https://doi.org/10.1016/j.jhazmat.2023.132016
Hill MA, Meininger GA (2012) Arteriolar vascular smooth muscle cells: mechanotransducers in a complex environment. Int J Biochem Cell B 44:1505–1510. https://doi.org/10.1016/j.biocel.2012.05.021
Yu XS, Hu LZ, He H et al (2019) Y-shaped DNA-mediated hybrid nanoflowers as efficient gene carriers for fluorescence imaging of tumor-related mRNA in living cells. Anal Chim Acta 1057:114–122. https://doi.org/10.1016/j.aca.2018.12.062
Wu QL, Liu ZJ, Su L et al (2018) Sticky-flares for monitoring of human telomerase RNA in living cells. Nanoscale 10:9386–9392. https://doi.org/10.1039/C8NR01260A
Nielsen PE, Egholm M, Berg RH, Buchardt O (1991) Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 254:1497–1500. https://doi.org/10.1126/science.1962210
Funding
This work was supported by the National Natural Science Foundation of China (21703255), Anhui Province Science Fund for Outstanding Young Scholars (2208085Y27), Scientific Research Foundation for the Returned Overseas Chinese Scholars of Anhui Province-Key Projects (2021LCX003), Excellent Young Talents Fund Program of Higher Education Institutions of Anhui Province (gxyqZD2021102), Research Fund of Anhui Institute of Translational Medicine (2021zhyx-C17), Research Fund for the Doctoral Program of Anhui Medical University (XJ201808), and Research Fund for Scientific Research Level Improvement Plan of Anhui Medical University (2020xkjT001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval
This is an observational study. The Ethics Committee of Anhui Medical University has confirmed that no ethical approval is required.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dong, F., Yan, W., Qu, M. et al. Strand displacement–triggered FRET nanoprobe tracking TK1 mRNA in living cells for ratiometric fluorimetry of nucleic acid biomarker. Microchim Acta 191, 390 (2024). https://doi.org/10.1007/s00604-024-06453-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-024-06453-7