Abstract
Background
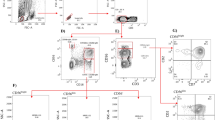
Natural killer (NK) cells play an important role in immune responses to virus infection. The cell population consists of CD56bright (bright-subset) and CD56dim (dim-subset) subsets that possess armed functions of cytokine production and cytolysis, respectively. How these subsets are involved in chronic hepatitis C virus infection (CHC) remains obscure.
Methods
We investigated the frequency, phenotype, and cytokine response of these subsets in blood from CHC patients and healthy subjects (HS).
Results
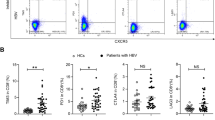
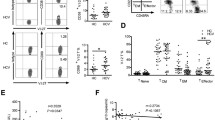
Dim-subset, but not bright-subset, showed lower frequency in the patients than in HS. Bright-subset from the patients more frequently expressed the NKG2A/CD94 inhibitory receptor than that from HS, while both subsets from the patients expressed lower levels of the NKG2D activating receptor. Both subsets from the patients displayed a significantly higher level of the signal transducer and activator of transcription (STAT) 1, compared with the HS. Upon stimulation with interferon-α, bright-subset activated less STAT4, required for interferon-γ production, and dim-subset activated more STAT1, required for cytolysis, in the patients than in HS.
Conclusions
These results indicate alterations of NK cell subsets in frequency, phenotype, and cytokine response in CHC, which might be associated with the immune pathogenesis of CHC.






Similar content being viewed by others
Abbreviations
- NK:
-
Natural killer
- IFN:
-
Interferon
- IL:
-
Interleukin
- HCV:
-
Hepatitis C virus
- CHC:
-
Chronic hepatitis C virus infection
- CHB:
-
Chronic hepatitis B virus infection
- PBMC:
-
Peripheral blood mononuclear cell
- STAT:
-
Signal transducer and activator of transcription
- pSTAT:
-
Phosphorylated-signal transducer and activator of transcription
- HS:
-
Healthy subjects
- ISG:
-
Interferon-stimulated gene
- MICA:
-
Major histocompatibility complex class I-related chain A
References
Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220.
Farrar MA, Schreiber RD. The molecular cell biology of interferon-gamma and its receptor. Annu Rev Immunol. 1993;11:571–611.
Lee SH, Miyagi T, Biron CA. Kee** NK cells in highly regulated antiviral warfare. Trends Immunol. 2007;28:252–9.
Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–40.
Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–9.
Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305.
Kamal SM, Fouly AE, Kamel RR, Hockenjos B, Al Tawil A, Khalifa KE, et al. Peginterferon alfa-2b therapy in acute hepatitis C: impact of onset of therapy on sustained virologic response. Gastroenterology. 2006;130:632–8.
Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–65.
Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL Jr, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–82.
Santantonio T, Fasano M, Sinisi E, Guastadisegni A, Casalino C, Mazzola M, et al. Efficacy of a 24-week course of PEG-interferon alpha-2b monotherapy in patients with acute hepatitis C after failure of spontaneous clearance. J Hepatol. 2005;42:329–33.
Meier UC, Owen RE, Taylor E, Worth A, Naoumov N, Willberg C, et al. Shared alterations in NK cell frequency, phenotype, and function in chronic human immunodeficiency virus and hepatitis C virus infections. J Virol. 2005;79:12365–74.
Morishima C, Paschal DM, Wang CC, Yoshihara CS, Wood BL, Yeo AE, et al. Decreased NK cell frequency in chronic hepatitis C does not affect ex vivo cytolytic killing. Hepatology. 2006;43:573–80.
Nattermann J, Feldmann G, Ahlenstiel G, Langhans B, Sauerbruch T, Spengler U. Surface expression and cytolytic function of natural killer cell receptors is altered in chronic hepatitis C. Gut. 2006;55:869–77.
Golden-Mason L, Madrigal-Estebas L, McGrath E, Conroy MJ, Ryan EJ, Hegarty JE, et al. Altered natural killer cell subset distributions in resolved and persistent hepatitis C virus infection following single source exposure. Gut. 2008;57:1121–8.
Bonorino P, Ramzan M, Camous X, Dufeu-Duchesne T, Thélu MA, Sturm N, et al. Fine characterization of intrahepatic NK cells expressing natural killer receptors in chronic hepatitis B and C. J Hepatol. 2009;51:458–67.
Oliviero B, Varchetta S, Paudice E, Michelone G, Zaramella M, Mavilio D, et al. Natural killer cell functional dichotomy in chronic hepatitis B and chronic hepatitis C virus infections. Gastroenterology. 2009;137:1151–60, 1160.e1–7.
Takehara T, Hayashi N. Natural killer cells in hepatitis C virus infection: from innate immunity to adaptive immunity. Clin Gastroenterol Hepatol. 2005;3:S78–81.
Golden-Mason L, Rosen HR. Natural killer cells: primary target for hepatitis C virus immune evasion strategies? Liver Transpl. 2006;12:363–72.
Szabo G, Chang S, Dolganiuc A. Altered innate immunity in chronic hepatitis C infection: cause or effect? Hepatology. 2007;46:1279–90.
Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289–93.
**ushi M, Takehara T, Tatsumi T, Kanto T, Miyagi T, Suzuki T, et al. Negative regulation of NK cell activities by inhibitory receptor CD94/NKG2A leads to altered NK cell-induced modulation of dendritic cell functions in chronic hepatitis C virus infection. J Immunol. 2004;173:6072–81.
Miyagi T, Gil MP, Wang X, Louten J, Chu WM, Biron CA. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J Exp Med. 2007;204:2383–96.
Miyagi T, Lee SH, Biron CA. Intracellular staining for analysis of the expression and phosphorylation of signal transducers and activators of transcription (STATs) in NK cells. Methods Mol Biol. 2010;612:159–75.
Miyagi T, Takehara T, Nishio K, Shimizu S, Kohga K, Li W, et al. Altered interferon-alpha-signaling in natural killer cells from patients with chronic hepatitis C virus infection. J Hepatol. 2010;53:424–30.
Uzel G, Frucht DM, Fleisher TA, Holland SM. Detection of intracellular phosphorylated STAT-4 by flow cytometry. Clin Immunol. 2001;100:270–6.
Krutzik PO, Clutter MR, Nolan GP. Coordinate analysis of murine immune cell surface markers and intracellular phosphoproteins by flow cytometry. J Immunol. 2005;175:2357–65.
Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–8.
Ji X, Cheung R, Cooper S, Li Q, Greenberg HB, He XS. Interferon alfa regulated gene expression in patients initiating interferon treatment for chronic hepatitis C. Hepatology. 2003;37:610–21.
Pfeffer LM, Madey MA, Riely CA, Fleckenstein JF. The induction of type I interferon production in hepatitis C-infected patients. J Interferon Cytokine Res. 2009;29:299–306.
Dolganiuc A, Norkina O, Kodys K, Catalano D, Bakis G, Marshall C, et al. Viral and host factors induce macrophage activation and loss of toll-like receptor tolerance in chronic HCV infection. Gastroenterology. 2007;133:1627–36.
Chen L, Borozan I, Sun J, Guindi M, Fischer S, Feld J, et al. Cell-type specific gene expression signature in liver underlies response to interferon therapy in chronic hepatitis C infection. Gastroenterology. 2010;138:1123–33.
Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci USA. 2008;105:7034–9.
Tateno M, Honda M, Kawamura T, Honda H, Kaneko S. Expression profiling of peripheral-blood mononuclear cells from patients with chronic hepatitis C undergoing interferon therapy. J Infect Dis. 2007;195:255–67.
Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Kaneko S. Different signaling pathways in the livers of patients with chronic hepatitis B or chronic hepatitis C. Hepatology. 2006;44:1122–38.
Amadei B, Urbani S, Cazaly A, Fisicaro P, Zerbini A, Ahmed P, et al. Activation of natural killer cells during acute infection with hepatitis C virus. Gastroenterology. 2010;138:1536–45.
Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE Jr. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci USA. 1996;93:7673–8.
Tanabe Y, Nishibori T, Su L, Arduini RM, Baker DP, David M. Cutting edge: role of STAT1, STAT3, and STAT5 in IFN-alpha beta responses in T lymphocytes. J Immunol. 2005;174:609–13.
García-Sastre A, Biron CA. Type 1 interferons and the virus–host relationship: a lesson in détente. Science. 2006;312:879–82.
Cho SS, Bacon CM, Sudarshan C, Rees RC, Finbloom D, Pine R, et al. Activation of STAT4 by IL-12 and IFN-alpha: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J Immunol. 1996;157:4781–9.
Matikainen S, Paananen A, Miettinen M, Kurimoto M, Timonen T, Julkunen I, et al. IFN-alpha and IL-18 synergistically enhance IFN-gamma production in human NK cells: differential regulation of Stat4 activation and IFN-gamma gene expression by IFN-alpha and IL-12. Eur J Immunol. 2001;31:2236–45.
Liang S, Wei H, Sun R, Tian Z. IFN alpha regulates NK cell cytotoxicity through STAT1 pathway. Cytokine. 2003;23:190–9.
Ahlenstiel G, Titerence RH, Koh C, Edlich B, Feld JJ, Rotman Y, et al. Natural killer cells are polarized toward cytotoxicity in chronic hepatitis C in an interferon-alfa-dependent manner. Gastroenterology. 2010;138:325–35.
Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, et al. Hepatic gene expression during treatment with peginterferon and ribavirin: identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–63.
Kohga K, Takehara T, Tatsumi T, Ohkawa K, Miyagi T, Hiramatsu N, et al. Serum levels of soluble major histocompatibility complex (MHC) class I-related chain A in patients with chronic liver diseases and changes during transcatheter arterial embolization for hepatocellular carcinoma. Cancer Sci. 2008;99:1643–9.
Acknowledgments
This work was supported by Grants-in-aid for Scientific Research (to T. Takehara and T. Miyagi) and by a grant for the Global Centers of Excellence Program (to T. Miyagi) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and a Grant-in-aid (to T. Takehara) from the Ministry of Health, Labour and Welfare of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
T. Miyagi and S. Shimizu contributed equally to this work and share the first authorship.
Rights and permissions
About this article
Cite this article
Miyagi, T., Shimizu, S., Tatsumi, T. et al. Differential alteration of CD56bright and CD56dim natural killer cells in frequency, phenotype, and cytokine response in chronic hepatitis C virus infection. J Gastroenterol 46, 1020–1030 (2011). https://doi.org/10.1007/s00535-011-0408-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-011-0408-8