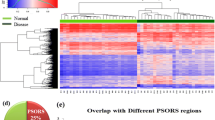
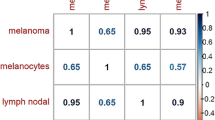
Abstract
Several studies have emphasized the role of DNA methylation in vitiligo. However, its profile in human skin of individuals with vitiligo remains unknown. Here, we aimed to study the DNA methylation profile of vitiligo using pairwise comparisons of lesions, peri-lesions, and healthy skin. We investigated DNA methylation levels in six lesional skin, six peri-lesional skin, and eight healthy skin samples using an Illumina 850 K methylation chip. We then integrated DNA methylation data with transcriptome data to identify differentially methylated and expressed genes (DMEGs) and analyzed their functional enrichment. Subsequently, we compared the methylation and transcriptome characteristics of all skin samples, and the related genes were further studied using scRNA-seq data. Finally, validation was performed using an external dataset. We observed more DNA hypomethylated sites in patients with vitiligo. Further integrated analysis identified 264 DMEGs that were mainly functionally enriched in cell division, pigmentation, circadian rhythm, fatty acid metabolism, peroxidase activity, synapse regulation, and extracellular matrix. In addition, in the peri-lesional skin, we found that methylation levels of 102 DMEGs differed prior to changes in their transcription levels and identified 16 key pre-DMEGs (ANLN, CDCA3, CENPA, DEPDC1, ECT2, DEPDC1B, HMMR, KIF18A, KIF18B, TTK, KIF23, DCT, EDNRB, MITF, OCA2, and TYRP1). Single-cell RNA analysis showed that these genes were associated with cycling keratinocytes and melanocytes. Further analysis of cellular communication indicated the involvement of the extracellular matrix. The expression of related genes was verified using an external dataset. To the best of our knowledge, this is the first study to report a comprehensive DNA methylation profile of clinical vitiligo and peri-lesional skin. These findings would contribute to future research on the pathogenesis of vitiligo and potential therapeutic strategies.






Similar content being viewed by others
Availability of data and materials
They are available from the corresponding author upon reasonable request.
References
Abedin M, King N (2010) Diverse evolutionary paths to cell adhesion. Trends Cell Biol 20(12):734–742
Bastonini E, Kovacs D, Picardo M (2016) Skin pigmentation and pigmentary disorders: focus on epidermal/dermal cross-talk. Ann Dermatol 28(3):279–289
Bergqvist C, Ezzedine K (2020) Vitiligo: a review. Dermatology (Basel, Switzerland) 236(6):571–592
Bibeau K, Pandya AG, Ezzedine K, Jones H, Gao J, Lindley A et al (2022) Vitiligo prevalence and quality of life among adults in Europe, Japan and the USA. J Eur Acad Dermatol Venereol 36(10):1831–1844
Brandner JM, Haass NK (2013) Melanoma’s connections to the tumour microenvironment. Pathology 45(5):443–452
Chen J, Li S, Li C (2021) Mechanisms of melanocyte death in vitiligo. Med Res Rev 41(2):1138–1166
Chen KD, Huang YH, Guo MM, Chang LS, Chu CH, Bu LF et al (2022) DNA methylation array identifies golli-MBP as a biomarker for disease severity in childhood atopic dermatitis. J Invest Dermatol 142(1):104–113
Delić D, Wolk K, Schmid R, Gabrielyan O, Christou D, Rieber K et al (2020) Integrated microRNA/mRNA expression profiling of the skin of psoriasis patients. J Dermatol Sci 97(1):9–20
Ezzedine K, Eleftheriadou V, Whitton M, van Geel N (2015) Vitiligo. Lancet (Lond, Engl) 386(9988):74–84
Ezzedine K, Eleftheriadou V, Jones H, Bibeau K, Kuo FI, Sturm D et al (2021) Psychosocial effects of vitiligo: a systematic literature Review. Am J Clin Dermatol 22(6):757–774
Farag AGA, Badr EAE, El-Shafey AES, Elshaib ME (2022) Fatty acid-binding protein 4 circulating levels in non-segmental vitiligo. Anais Bras De Dermatol 97(1):28–36
Frisoli ML, Essien K, Harris JE (2020) Vitiligo: mechanisms of pathogenesis and treatment. Annu Rev Immunol 26(38):621–648
Gellatly KJ, Strassner JP, Essien K, Refat MA, Murphy RL, Coffin-Schmitt A et al (2021) scRNA-seq of human vitiligo reveals complex networks of subclinical immune activation and a role for CCR5 in T(reg) function. Sci Transl Med 13(610):eabd8995
Hirobe T, Furuya R, Akiu S, Ifuku O, Fukuda M (2002) Keratinocytes control the proliferation and differentiation of cultured epidermal melanocytes from ultraviolet radiation B-induced pigmented spots in the dorsal skin of hairless mice. Pigment Cell Res 15(5):391–399
Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J et al (2010) Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res 20(2):170–179
Köhler F, Rodríguez-Paredes M (2020) DNA methylation in epidermal differentiation, aging, and cancer. J Invest Dermatol 140(1):38–47
Le Poole IC, van den Wijngaard RM, Westerhof W, Das PK (1997) Tenascin is overexpressed in vitiligo lesional skin and inhibits melanocyte adhesion. Br J Dermatol 137(2):171–178
Lei W, Luo Y, Lei W, Luo Y, Yan K, Zhao S et al (2009) Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand J Rheumatol 38(5):369–374
López S, Alonso S, García de Galdeano A, Smith-Zubiaga I (2015) Melanocytes from dark and light skin respond differently after ultraviolet B irradiation: effect of keratinocyte-conditioned medium. Photodermatol Photoimmunol Photomed 31(3):149–158
Mei L, Ying L, Wang H, Xu G, Ye X, Yang G (2022) (1)H NMR-based metabolomics of skin squamous cell carcinoma and peri-tumoral region tissues. J Pharm Biomed Anal 1(212):114643
Michel G, Tonon T, Scornet D, Cock JM, Kloareg B (2010) The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol 188(1):82–97
Mozzanica N, Frigerio U, Negri M, Tadini G, Villa ML, Mantovani M et al (1989) Circadian rhythm of natural killer cell activity in vitiligo. J Am Acad Dermatol 20(4):591–596
Mozzanica N, Frigerio U, Finzi AF, Cattaneo A, Negri M, Scaglione F et al (1990) T cell subpopulations in vitiligo: a chronobiologic study. J Am Acad Dermatol 22(2 Pt 1):223–230
Mozzanica N, Villa ML, Foppa S, Vignati G, Cattaneo A, Diotti R et al (1992) Plasma alpha-melanocyte-stimulating hormone, beta-endorphin, met-enkephalin, and natural killer cell activity in vitiligo. J Am Acad Dermatol 26(5 Pt 1):693–700
Pu Y, Chen X, Chen Y, Zhang L, Chen J, Zhang Y et al (2021) Transcriptome and differential methylation integration analysis identified important differential methylation annotation genes and functional epigenetic modules related to vitiligo. Front Immunol 12:587440
Rani S, Pervaiz N, Parsad D, Kumar R (2023) Differential expression of extracellular matrix proteins in the lesional skin of vitiligo patients. Arch Dermatol Res. https://doi.org/10.1007/s00403-023-02628-z
Rasheed H, El-Komy M, Hegazy RA, Gawdat HI, AlOrbani AM, Shaker OG (2016) Expression of sirtuins 1, 6, tumor necrosis factor, and interferon-γ in psoriatic patients. Int J Immunopathol Pharmacol 29(4):764–768
Regazzetti C, Joly F, Marty C, Rivier M, Mehul B, Reiniche P et al (2015) Transcriptional analysis of vitiligo skin reveals the alteration of WNT pathway: a promising target for repigmenting vitiligo patients. J Invest Dermatol 135(12):3105–3114
Roberson ED, Liu Y, Ryan C, Joyce CE, Duan S, Cao L et al (2012) A subset of methylated CpG sites differentiate psoriatic from normal skin. J Invest Dermatol 132(3 Pt 1):583–592
Shiu J, Zhang L, Lentsch G, Flesher JL, ** S, Polleys C et al (2022) Multimodal analyses of vitiligo skin identify tissue characteristics of stable disease. JCI Insight. https://doi.org/10.1172/jci.insight.154585
Singh A, Gotherwal V, Junni P, Vijayan V, Tiwari M, Ganju P et al (2017) Map** architectural and transcriptional alterations in non-lesional and lesional epidermis in vitiligo. Sci Rep 7(1):9860
Verma D, Ekman AK, BivikEding C, Enerbäck C (2018) Genome-wide DNA methylation profiling identifies differential methylation in uninvolved psoriatic epidermis. J Invest Dermatol 138(5):1088–1093
Xuan Y, Yang Y, **ang L, Zhang C (2022) The role of oxidative stress in the pathogenesis of vitiligo: a culprit for melanocyte death. Oxid Med Cell Longev 2022:8498472
Ye Z, Chen J, Du P, Ni Q, Li B, Zhang Z et al (2022) Metabolomics signature and potential application of serum polyunsaturated fatty acids metabolism in patients with vitiligo. Front Immunol 13:839167
Zhao M, Gao F, Wu X, Tang J, Lu Q (2010) Abnormal DNA methylation in peripheral blood mononuclear cells from patients with vitiligo. Br J Dermatol 163(4):736–742
Zhao M, Huang W, Zhang Q, Gao F, Wang L, Zhang G et al (2012) Aberrant epigenetic modifications in peripheral blood mononuclear cells from patients with pemphigus vulgaris. Br J Dermatol 167(3):523–531
Zhao M, Liang G, Wu X, Wang S, Zhang P, Su Y et al (2012) Abnormal epigenetic modifications in peripheral blood mononuclear cells from patients with alopecia areata. Br J Dermatol 166(2):226–273
Zhou F, Shen C, Xu J, Gao J, Zheng X, Ko R et al (2016) Epigenome-wide association data implicates DNA methylation-mediated genetic risk in psoriasis. Clin Epigenetics 8:131
Acknowledgements
We are very grateful to the patients and volunteers involved in this study.
Funding
The National Natural Science Foundation of China (82073462). Chongqing Natural Science Foundation (CSTB2023NSCQ- MSX0664 and CSTB2023NSCQ- MSX0075).
Author information
Authors and Affiliations
Contributions
Conceptualization, LL and YX. Data collection and processing, YL, XP, and YC. Interpretation of data, YH. Software, TC, XS, and JZ. LL is responsible for writing the initial article. Revision and finalization, YP and JC. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Conflict of interest
None declared.
Ethical approval and consent to participate
For patient samples, written informed consent was obtained from each patient and the study was approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (No.:2023-126). The study was performed in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
439_2023_2630_MOESM1_ESM.tif
Supplementary file1 Figure S1. (A) Venn plots of the overlap of DEGs in the pairwise comparisons of L, PL, and HC. Gene ontology enrichment of DEGs of (B) L vs. HC, (C) PL vs. HC, and (D) L vs. PL. L, lesion; HC, healthy control; PL, peri-lesion. (TIF 2120 kb)
439_2023_2630_MOESM2_ESM.tif
Supplementary file2 Figure S2. (A) PPI network of 264 DMEGs. (B) PPI network of 102 pre-DMEGs. PPI, Protein-Protein Interaction; DMEGs, differentially methylated and expressed genes. (TIF 3803 kb)
439_2023_2630_MOESM3_ESM.tif
Supplementary file3 Figure S3. LASSO logistic regression algorithm. (A) LASSO coefficient profiles. (B) Cross-validation for tuning parameter selection in the LASSO model. (TIF 1183 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, L., Xue, Y., Li, Y. et al. Genome-wide DNA methylation of lesional and peri-lesional skin in vitiligo: a comparative and integrated analysis of multi-omics in Chinese population. Hum. Genet. 143, 137–149 (2024). https://doi.org/10.1007/s00439-023-02630-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-023-02630-5