Abstract
Purpose
Breast cancer patients typically have decent prognoses, with a 5-year survival rate of more than 90%, but when the disease metastases to lymph node or distant, the prognosis drastically declines. Therefore, it is essential for future treatment and patient survival to quickly and accurately identify tumor metastasis in patients. An artificial intelligence system was developed to recognize lymph node and distant tumor metastases on whole-slide images (WSIs) of primary breast cancer.
Methods
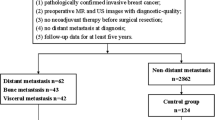
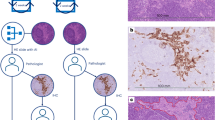
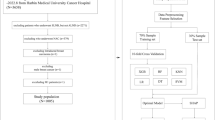
In this study, a total of 832 WSIs from 520 patients without tumor metastases and 312 patients with breast cancer metastases (including lymph node, bone, lung, liver, and other) were gathered. Based on the WSIs were randomly divided into the training and testing cohorts, a brand-new artificial intelligence system called MEAI was built to identify lymph node and distant metastases in primary breast cancer.
Results
The final AI system attained an area under the receiver operating characteristic curve of 0.934 in a test set of 187 patients. In addition, the potential for AI system to increase the precision, consistency, and effectiveness of tumor metastasis detection in patients with breast cancer was highlighted by the AI’s achievement of an AUROC higher than the average of six board-certified pathologists (AUROC 0.811) in a retrospective pathologist evaluation.
Conclusion
The proposed MEAI system can provide a non-invasive approach to assess the metastatic probability of patients with primary breast cancer.








Similar content being viewed by others
Data Availability
Data are available in the article. All other data can be provided upon reasonable request to the corresponding authors.
Code availability
The code of the proposed method can be provided upon reasonable request to the corresponding authors.
References
Afkari H, Makrufardi F, Hidayat B, Budiawan H, Kartamihardja AHS (2021) Correlation between ER, PR, HER-2, and Ki-67 with the risk of bone metastases detected by bone scintigraphy in breast cancer patients: a cross sectional study. Ann Med Surg 67:102532
Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ (2017) Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging 30(4):449–459
Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK et al (2017) The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA: Cancer J Clin 67(2):93–99
Bejnordi BE, Veta M, Van Diest PJ, Van Ginneken B, Karssemeijer N, Litjens G et al (2017) Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318(22):2199–2210
Chen Z, Chi Z, Fu H, Feng D (2013) Multi-instance multi-label image classification: a neural approach. Neurocomputing 99:298–306
Cserni G, Amendoeira I, Apostolikas N, Bellocq J, Bianchi S, Boecker W et al (2004) Discrepancies in current practice of pathological evaluation of sentinel lymph nodes in breast cancer. Results of a questionnaire based survey by the European Working Group for Breast Screening Pathology. J Clin Pathol 57(7):695–701
Diao JA, Wang JK, Chui WF, Mountain V, Gullapally SC, Srinivasan R et al (2021) Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat Commun 12(1):1–15
Dietterich TG, Lathrop RH, Lozano-Pérez T (1997) Solving the multiple instance problem with axis-parallel rectangles. Artif Intell 89(1–2):31–71
Fan K, Wen S, Deng Z (2019) Deep learning for detecting breast cancer metastases on WSI. In: Chen YW (ed) Innovation in medicine and healthcare systems, and multimedia. Springer, Berlin, pp 137–145
Feng R, Liu X, Chen J, Chen DZ, Gao H, Wu J (2020) A deep learning approach for colonoscopy pathology WSI analysis: accurate segmentation and classification. IEEE J Biomed Health Inform 25(10):3700–3708
Fu Y, Jung AW, Torne RV, Gonzalez S, Vöhringer H, Shmatko A et al (2020) Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat Cancer 1(8):800–810
Greenwald NF, Miller G, Moen E, Kong A, Kagel A, Dougherty T et al (2022) Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat Biotechnol 40(4):555–565
Guo Z, Li X, Huang H, Guo N, Li Q (2019) Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans Radiat Plasma Med Sci 3(2):162–169
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Howlader N, Noone A, Krapcho M, Miller D, Bishop K, Altekruse S et al (2016) SEER cancer statistics review, 1975–2013. National Cancer Institute, USA, pp 1992–2013
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Huang SC, Chen CC, Lan J, Hsieh TY, Chuang HC, Chien MY et al (2022a) Deep neural network trained on gigapixel images improves lymph node metastasis detection in clinical settings. Nat Commun 13(1):1–14
Huang L, Zhang XO, Rozen EJ, Sun X, Sallis B, Verdejo-Torres O et al (2022b) PRMT5 activates AKT via methylation to promote tumor metastasis. Nat Commun 13(1):1–16
Ilse M, Tomczak J, Welling M (2018) Attention-based deep multiple instance learning. In: International conference on machine learning. PMLR, pp 2127–2136
Jaber MI, Song B, Taylor C, Vaske CJ, Benz SC, Rabizadeh S et al (2020) A deep learning image-based intrinsic molecular subtype classifier of breast tumors reveals tumor heterogeneity that may affect survival. Breast Cancer Res 22(1):1–10
Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH et al (2019) Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 25(7):1054–1056
Kim JH, Choo W, Jeong H, Song HO (2021) Co-mixup: saliency guided joint mixup with supermodular diversity. ar**v preprint. ar**v:2102.03065
Krag DN, Anderson SJ, Julian TB, Brown AM, Harlow SP, Ashikaga T et al (2007) Technical outcomes of sentinel-lymph-node resection and conventional axillary-lymph-node dissection in patients with clinically node-negative breast cancer: results from the NSABP B-32 randomised phase III trial. Lancet Oncol 8(10):881–888
Kundu A, Yin X, Fathi A, Ross D, Brewington B, Funkhouser T et al (2020) Virtual multi-view fusion for 3d semantic segmentation. In: European conference on computer vision. Springer, Berlin, pp 518–535
Lin CJ, Jeng SY (2020) Optimization of deep learning network parameters using uniform experimental design for breast cancer histopathological image classification. Diagnostics 10(9):662
Liu C, Ding J, Spuhler K, Gao Y, Serrano Sosa M, Moriarty M et al (2019) Preoperative prediction of sentinel lymph node metastasis in breast cancer by radiomic signatures from dynamic contrast-enhanced MRI. J Magn Reson Imaging 49(1):131–140
Liu F, Hardiman T, Wu K, Quist J, Gazinska P, Ng T et al (2021) Systemic immune reaction in axillary lymph nodes adds to tumor-infiltrating lymphocytes in triple-negative breast cancer prognostication. NPJ Breast Cancer 7(1):1–10
Lv T, Wu Y, Wang Y, Liu Y, Li L, Deng C et al (2022) A hybrid hemodynamic knowledge-powered and feature reconstruction-guided scheme for breast cancer segmentation based on DCE-MRI. Med Image Anal 82:102572
Naik N, Madani A, Esteva A, Keskar NS, Press MF, Ruderman D et al (2020) Deep learning-enabled breast cancer hormonal receptor status determination from base-level H &E stains. Nat Commun 11(1):1–8
Orr ER, Ballantyne M, Gonzalez A, Jack SM (2020) Visual elicitation: methods for enhancing the quality and depth of interview data in applied qualitative health research. Adv Nurs Sci 43(3):202–213
Qu H, Zhou M, Yan Z, Wang H, Rustgi VK, Zhang S et al (2021) Genetic mutation and biological pathway prediction based on whole slide images in breast carcinoma using deep learning. NPJ Precis Oncol 5(1):1–11
Shen Y, Shamout FE, Oliver JR, Witowski J, Kannan K, Park J et al (2021) Artificial intelligence system reduces false-positive findings in the interpretation of breast ultrasound exams. Nat Commun 12(1):1–13
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. ar**v preprint. ar**v:1409.1556
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Tian Q, Wang Y, Guo H, **e G, Li J, Zhang M et al (2017) Recent perspectives of management of breast cancer metastasis—an update. J BUON 22(2):295–300
Vahadane A, Peng T, Sethi A, Albarqouni S, Wang L, Baust M et al (2016) Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging 35(8):1962–1971
Wang D, Khosla A, Gargeya R, Irshad H, Beck AH (2016) Deep learning for identifying metastatic breast cancer. ar**v preprint. ar**v:1606.05718
Wilke LG, McCall LM, Posther KE, Whitworth PW, Reintgen DS, Leitch AM et al (2006) Surgical complications associated with sentinel lymph node biopsy: results from a prospective international cooperative group trial. Ann Surg Oncol 13(4):491–500
Xu F, Zhu C, Tang W, Wang Y, Zhang Y, Li J et al (2021) Predicting axillary lymph node metastasis in early breast cancer using deep learning on primary tumor biopsy slides. Front Oncol 11:759007
Zhang Y, Sidibé D, Morel O, Mériaudeau F (2021a) Deep multimodal fusion for semantic image segmentation: a survey. Image Vis Comput 105:104042
Zhang J, Wei Q, Dong D, Ren L (2021b) The role of TPS, CA125, CA15-3 and CEA in prediction of distant metastasis of breast cancer. Clin Chim Acta 523:19–25
Acknowledgements
We acknowledge the pathologists of the Affiliated Hospital of Jiangnan University for assistance with pathologic diagnosis.
Funding
This work was supported in part by the National Key R &D Program of China under Grants 2021YFE0203700 and 2018YFA0701700, the Postgraduate Research and Practice Innovation Program of Jiangsu Province SJCX22_1106, and was supported by National Natural Science Foundation of China grants 61602007, U21A20521 and 61731008, Zhejiang Provincial Natural Science Foundation of China (LZ15F010001), the University of Macau (grant numbers: FHS-CRDA-029-002-2017 and MYRG2018-00071-FHS) and the Science and Technology Development Fund, Macau SAR (File no.0004/2019/AFJ and 0011/2019/AKP).
Author information
Authors and Affiliations
Contributions
XP is the guarantor of this study. XP, CJ, and LL conceived and supervised this study. HS, TL, KM, and YL collected the digital slides and performed the preprocessing. JF and LZ designed and conducted the experiments. JF, XP, CJ, and LL performed statistical analyses of the results. JF and LZ drafted the manuscript. All authors approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors of this manuscript declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fan, J., Zhang, L., Lv, T. et al. MEAI: an artificial intelligence platform for predicting distant and lymph node metastases directly from primary breast cancer. J Cancer Res Clin Oncol 149, 9229–9241 (2023). https://doi.org/10.1007/s00432-023-04787-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-04787-y