Abstract
Objective
To use weighted gene correlation network analysis (WGCNA) and machine learning algorithm to predict classification of early pulmonary nodes with public databases.
Methods
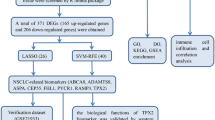
The expression data and clinical data of lung cancer patients were firstly extracted from public database (GTEx and TCGA) to study the differentially expressed genes (DEGs) of lung adenocarcinoma (LUAD). The intersection of three R packages (Dseq2, Limma, EdgeR) methods were selected as candidate DEGs for further study. WGCNA was used to obtain relevant modules and key genes of lung cancer classification, GO and KEGG enrichment analysis was performed. The model was built using two machine learning methods, Least Absolute Shrinkage and Selection Operator (LASSO) regression and tumor classification was also predicted with extreme Gradient Boosting (XGBoost) algorithm.
Results
DEGs analysis revealed that there were 1306 LUAD genes. WGCNA module analysis showed that a total of 116 genes were significantly related to classification, and module genes were mainly related to 14 KEGG pathways. The machine learning algorithm identified 10 target genes by LASSO regression analysis of differential genes, and 18 genes were identified by XGBoost model. A total of 6 genes were found from the intersection of the above methods as classification signatures of early pulmonary nodules, including “HMGB3” “ARHGAP6” “TCF21” “FCN3” “COL6A6” “GOLM1”.
Conclusion
Using DEGs analysis, WGCNA method and machine learning algorithm, six gene signatures related to early stage of LUAD, which can assist clinicians in disease classification prediction.






Similar content being viewed by others
References
Alaei S, Sadeghi B, Najafi A et al (2019) LncRNA and mRNA integration network reconstruction reveals novel key regulators in esophageal squamous-cell carcinoma. Genomics 111(1):76–89
Alizadeh AA, Eisen MB, Davis RE et al (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403(6769):503
Andrea F, Damian S, Sune F et al (2012) STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucl Acids Res 41(1):D808–D815
Aruna LML (2018) Overexpression of golgi membrane protein 1 promotes non-small-cell carcinoma aggressiveness by regulating the matrix metallopeptidase 13. Am J Cancer Res 8(3):551–565
Chen T, Tong H, Benesty M (2016) xgboost: Extreme Gradient Boosting
Dibley MJ, Staehling N, Nieburg P et al (1987) Interpretation of Z-score anthropometric indicators derived from the international growth reference. Am J Clin Nutr 46(5):749–762
Goldberg SI, Niemierko A, Turchin A (2008) Analysis of data errors in clinical research databases. In: AMIA symposium. American Medical Informatics Association
Guo X, Wei Y, Zhe W et al (2018) LncRNA LINC00163 upregulation suppresses lung cancer development though transcriptionally increasing TCF21 expression. Am J Cancer Res 8(12):2494–2506
Guo C, Gao YY, Ju QQ et al (2021) The landscape of gene co-expression modules correlating with prognostic genetic abnormalities. AML J Transl Med. https://doi.org/10.1186/s12967-021-02914-2
Juarez-Flores A, Zamudio GS, José MV (2021) Novel gene signatures for stage classification of the squamous cell carcinoma of the lung. Sci Rep 11(4835):1–10
Keen JC, Moore HM (2015) The Genotype-Tissue Expression (GTEx) Project: linking clinical data with molecular analysis to advance personalized medicine. J Personal Med 5(1):22–29
Kim A, Sun ML, Kim JH et al (2021) Integrative genomic and transcriptomic analyses of tumor suppressor genes and their role on tumor microenvironment and immunity in lung squamous cell carcinoma. Front Immunol 12:598671
Kolde R (2015) pheatmap: Pretty Heatmaps
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinf 9(1):559
Liu J, Wang L, Li X (2018) HMGB3 promotes the proliferation and metastasis of glioblastoma and is negatively regulated by miR-200b-3p and miR-200c-3p. Cell Biochem Funct 36(7):357–365
Love M, Anders S, Huber W (2014) Differential analysis of count data—the deseq2 package
Lu M, Wu K, Trudeau S et al (2020) A genomic signature for accurate classification and prediction of clinical outcomes in cancer patients treated with immune checkpoint blockade immunotherapy. Sci Rep. https://doi.org/10.1038/s41598-020-77653-3
Ma Y, Qiu M, Guo H et al (2021) Comprehensive analysis of the immune and prognostic implication of COL6A6 in lung adenocarcinoma. Front Oncol 11:235
Mclendon R, Friedman A, Bigner D, Meir E, Meir EV, Brat D et al (2008) The Cancer Genome Atlas (TCGA), comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068
Raghav PK, Bhardwaj R, Raghava GPS (2019) Machine learning based identification of stem cell genes involved in stemness. J Cell Sci Ther 3:25–26
Ran LV, Liu Y, Wang PR (2017) Establishment of HPA gene database for platelet donors in **ngtai area of Hebei. J Clin Transfusion Lab Med
Reiner A, Yekutieli D, Benjamini Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19(3):368–375
Schelldorfer J, Meier L, Bühlmann P (2011) GLMMLasso: an algorithm for high-dimensional generalized linear mixed models using l(1)-penalization. J Comput Graph Stat 23(2):460–477
Smyth GK, Ritchie M, Thorne N, et al (2010) Limma: linear models for microarray data. In: Bioinformatics & computational biology solutions using R & bioconductor
Tang Z, Li C, Kang B et al (2017) (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucl Acids Res 1:W98–W102
Tinteren HV, Hoekstra OS, Smit EF et al (2002) Effectiveness of positron emission tomography in the preoperative assessment of patients with suspected non-small-cell lung cancer: the PLUS multicentre randomised trial. Lancet 359(9315):1388–1392
Vijay N, Poelstra JW, Künstner A, Wolf J (2012) Differential expression analysis—edgeR
Weinberg OK (2005) Aromatase inhibitors in human lung cancer therapy. Can Res 65(24):11287
Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R et al (2007) Characterizing the cancer genome in lung adenocarcinoma. Nature 450(7171):893–898
Wickham H (2009) Ggplot2: elegant graphics for data analysis. Springer Publishing Company, Incorporated
Xue K, Zheng H, Qian X et al (2021) Identification of key mRNAs as prediction models for early metastasis of pancreatic cancer based on LASSO. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2021.701039
Yin YJ et al (2016) Inhibitory effects of Arhgap6 on cervical carcinoma cells. Tumour Biol 37(2):1411–1425
Yu G, Wang LG, Han Y et al (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J Integr Biol 16(5):284–287
Ywh A, Clt A, Eycb C et al (2021) A risk prediction model of gene signatures in ovarian cancer through bagging of GA-XGBoost models. J Adv Res 30:113–122
Funding
This work was supported by Chongqing medical scientific research project (Joint project of Chongqing Health Commision and Science and Technology Bureau) under Grant No.2022DBXM005.
Author information
Authors and Affiliations
Contributions
FJ and LR conceived of the presented idea. GL developed the theory and performed the computations. MY verified the analytical methods. All authors discussed the results and contributed to the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
There were no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, G., Yang, M., Ran, L. et al. Classification prediction of early pulmonary nodes based on weighted gene correlation network analysis and machine learning. J Cancer Res Clin Oncol 149, 3915–3924 (2023). https://doi.org/10.1007/s00432-022-04312-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-04312-7