Abstract
Main conclusion
Natural selection influenced adaptive divergence between Cereus fernambucensis and Cereus insularis, revealing key genes governing abiotic stress responses and supporting neoteny in C. insularis.
Abstract
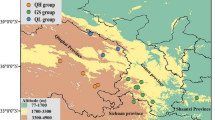
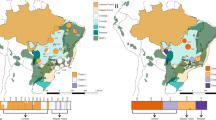
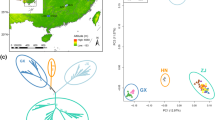
Uncovering the molecular mechanisms driving adaptive divergence in traits related to habitat adaptation remains a central challenge. In this study, we focused on the cactus clade, which includes Cereus sericifer F.Ritter, Cereus fernambucensis Lem., and Cereus insularis Hemsley. These allopatric species inhabit distinct relatively drier regions within the Brazilian Atlantic Forest, each facing unique abiotic conditions. We leveraged whole transcriptome data and abiotic variables datasets to explore lineage-specific and environment-specific adaptations in these species. Employing comparative phylogenetic methods, we identified genes under positive selection (PSG) and examined their association with non-synonymous genetic variants and abiotic attributes through a PhyloGWAS approach. Our analysis unveiled signatures of selection in all studied lineages, with C. fernambucensis northern populations and C. insularis showing the most PSGs. These PSGs predominantly govern abiotic stress regulation, encompassing heat tolerance, UV stress response, and soil salinity adaptation. Our exclusive observation of gene expression tied to early developmental stages in C. insularis supports the hypothesis of neoteny in this species. We also identified genes associated with abiotic variables in independent lineages, suggesting their role as environmental filters on genetic diversity. Overall, our findings suggest that natural selection played a pivotal role in the geographic range of these species in response to environmental and biogeographic transitions.




Similar content being viewed by others
Data availability
The datasets generated and analyzed in the present study are accessible through the NCBI repository under project number PRJNA1023027.
Abbreviations
- BAF:
-
Brazilian Atlantic Forest
- FNI:
-
Fernando de Noronha Island
- GO:
-
Gene Ontology
- GWAS:
-
Genome-Wide Association Studies
- PSG:
-
Positively selected gene
References
Adamczyk B, Adamczyk S, Smolander A, Kitunen V, Simon J (2018) Plant secondary metabolites—missing pieces in the soil organic matter puzzle of boreal forests. Soil Syst 2(1):2
Altesor A, Silva C, Ezcurra E (1994) Allometric neoteny and the evolution of succulence in cacti. Bot J Linn Soc 114(3):283–292
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Amaral DT, Bombonato JR, Andrade SCS, Moraes EM, Franco FF (2021a) The genome of a thorny species: comparative genomic analysis among South and North American Cactaceae. Planta 254(3):44
Amaral DT, Minhós-Yano I, Oliveira JVM, Romeiro-Brito M, Bonatelli IAS, Taylor NP et al (2021b) Tracking the xeric biomes of South America: the spatiotemporal diversification of mandacaru cactus. J Biogeogr 48(12):3085–3103
Ang LH, Deng XW (1994) Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6(5):613–628
Aquino D, Moreno-Letelier A, González-Botello MA, Arias S (2021) The importance of environmental conditions in maintaining lineage identity in Epithelantha (Cactaceae). Ecol Evol 11(9):4520–4531
Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65:415–442
Blankenship RE, Tiede DM, Barber J, Brudvig GW, Fleming G, Ghirardi M et al (2011) Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement. Science 332(6031):805–809
Bombonato JR, Amaral DT, Silva GAR, Khan G, Moraes EM, Andrade SCS et al (2020) The potential of genome-wide RAD sequences for resolving rapid radiations: a case study in Cactaceae. Mol Phylogenet Evol 151:106896
Buendía-Monreal M, Gillmor CS (2018) The times they are a-changin’: heterochrony in plant development and evolution. Front Plant Sci 9:1349
Bulgakov VP, Wu HC, **n TL (2019) Coordination of ABA and chaperone signaling in plant stress responses. Trends Plant Sci 24(7):636–651
Carnaval AC, Hickerson MJ, Haddad CF, Rodrigues MT, Moritz C (2009) Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science 323(5915):785–789
Carnaval AC, Waltari E, Rodrigues MT, Rosauer D, VanDerWal J, Damasceno R et al (2014) Prediction of phylogeographic endemism in an environmentally complex biome. Proc R Soc B 281(1792):20141461
Ciezarek AG, Osborne OG, Shipley ON, Brooks EJ, Tracey SR, McAllister JD et al (2019) Phylotranscriptomic insights into the diversification of endothermic Thunnus tunas. Mol Biol Evol 36(1):84–96
Divi UK, Krishna P (2009) Brassinosteroids confer stress tolerance. In: Hirt H (ed) Plant stress biology: from genomics to systems biology. Wiley-VCH, New York, pp 119–135
Donoghue MJ, Edwards EJ (2014) Biome shifts and niche evolution in plants. Annu Rev Ecol Evol Syst 45:547–572
Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238
Eshel G, Araus V, Undurraga S, Soto DC, Moraga C, Montecinos A et al (2021) Plant ecological genomics at the limits of life in the Atacama Desert. Proc Natl Acad Sci USA 118(46):e2101177118
Feder JL, Egan SP, Nosil P (2012) The genomics of speciation-with-gene-flow. Trends Genet 28(7):342–350
Fourounjian P, Slovin J, Messing J (2021) Flowering and seed production across the Lemnaceae. Int J Mol Sci 22(5):2733
Franco FF, Jojima CL, Perez MF, Zappi DC, Taylor N, Moraes EM (2017) The xeric side of the Brazilian Atlantic Forest: the forces sha** phylogeographic structure of cacti. Ecol Evol 7(22):9281–9293
Franco FF, Silva FA, Khan G, Bonatelli IA, Amaral DT, Zappi DC et al (2022) Generalizations of genetic conservation principles in islands are not always likely: a case study from a Neotropical insular cactus. Bot J Linn Soc 199(1):210–227
Franco FF, Amaral DT, Bonatelli IA, Meek JB, Moraes EM, Zappi DC et al (2024) A historical step**-stone path for an island-colonizing cactus across a submerged “bridge” archipelago. Heredity. https://doi.org/10.1038/s41437-024-00683-4
Fukushima K, Pollock DD (2023) Detecting macroevolutionary genotype–phenotype associations using error-corrected rates of protein convergence. Nat Ecol Evol 7(1):155–170
García-Verdugo C, Monroy P, Pugnaire FI, Jura-Morawiec J, Moreira X, Flexas J (2020) Leaf functional traits and insular colonization: subtropical islands as a melting pot of trait diversity in a widespread plant lineage. J Biogeogr 47(11):2362–2376
García-Verdugo C, Douthe C, Francisco M, Ribas-Carbó M, Flexas J, Moreira X (2023) Does insular adaptation to subtropical conditions promote loss of plasticity over time? Perspect Plant Ecol Evol Syst 58:125713
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I et al (2011) Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol 29(7):644
Gray JC (1992) Cytochrome f: Structure, function and biosynthesis. Photosynth Res 34:359–374
Hafez M, Rashad M, Popov AI (2020) The biological correction of agro-photosynthesis of soil plant productivity. J Plant Nutr 43(19):2929–2980
Hogan JA, Castañeda-Moya E, Lamb-Wotton L, Troxler T, Baraloto C (2022) Water levels primarily drive variation in photosynthesis and nutrient use of scrub Red Mangroves in the southeastern Florida Everglades. Tree Physiol 42(4):797–814
Huarancca-Reyes T, Scartazza A, Pompeiano A, Guglielminetti L (2019) Physiological responses of Lepidium meyenii plants to ultraviolet-B irradiation challenge. BMC Plant Biol 19(1):186
Hudson RR (2002) Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18(2):337–338
Kavas M, Yıldırım K, Seçgin Z, Abdulla MF, Gökdemir G (2021) Genome-wide identification of the BURP domain-containing genes in Phaseolus vulgaris. Physiol Mol Biol Plants 27(9):1885–1902
Kim MC, Lee SH, Kim JK, Chun HJ, Choi MS, Chung WS et al (2002) Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein: isolation and characterization of a rice Mlo homologue. J Biol Chem 277(22):19304–19314
Krishna P (2003) Brassinosteroid-mediated stress responses. J Plant Growth Regul 22(4):289–297
Lacerda CFD, Oliveira EVD, Neves AL, Gheyi HR, Bezerra MA, Costa CA (2020) Morphophysiological responses and mechanisms of salt tolerance in four ornamental perennial species under tropical climate. Rev Bras Eng Agrícola Ambient 24:656–663
Leite YL, Costa LP, Loss AC, Rocha RG, Batalha-Filho H, Bastos AC et al (2016) Neotropical forest expansion during the last glacial period challenges refuge hypothesis. Proc Natl Acad Sci USA 113(4):1008–1013
Lemmon ZH, Park SJ, Jiang K, Van Eck J, Schatz MC, Lippman ZB (2016) The evolution of inflorescence diversity in the nightshades and heterochrony during meristem maturation. Genome Res 26(12):1676–1686
Li J, Ren L, Gao Z, Jiang M, Liu Y, Zhou L et al (2017) Combined transcriptomic and proteomic analysis constructs a new model for light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L). Plant Cell Environ 40(12):3069–3087
Logan ML, Montgomery CE, Boback SM, Reed RN, Campbell JA (2012) Divergence in morphology, but not habitat use, despite low genetic differentiation among insular populations of the lizard Anolis lemurinus in Honduras. J Tropic Ecol 28(2):215–222
Maccioni A, Canopoli L, Cubeddu V, Cucca E, Dessena S, Morittu S et al (2021) Gradients of salinity and plant community richness and diversity in two different Mediterranean coastal ecosystems in NW Sardinia. Biodivers Data J 9:e71247
Martínez-Zamora L, Castillejo N, Artés-Hernández F (2021) Postharvest UV-B and UV-C irradiation enhanced the biosynthesis of glucosinolates and isothiocyanates in Brassicaceae sprouts. Postharvest Biol Tech 181:111650
Menezes MO, Taylor NP, Zappi DC, Loiola MIB (2015) Spines and ribs of Pilosocereus arrabidae (Lem) Byles GD Rowley and allies (Cactaceae) Ecologic or genetic traits? Flora Morphol Distrib Funct Ecol 214:44–49
Milici VR, Dalui D, Mickley JG, Bagchi R (2020) Responses of plant–pathogen interactions to precipitation Implications for tropical tree richness in a changing world. J Ecol 108(5):1800–1809
Nonogaki H (2010) MicroRNA gene regulation cascades during early stages of plant development. Plant Cell Physiol 51(11):1840–1846
Pease JB, Rosenzweig BK (2018) Encoding data using biological principles the multisample variant format for phylogenomics and population genomics. IEEE ACM Trans Comput Biol Bioinform 15(4):1231–1238
Pease JB, Haak DC, Hahn MW, Moyle LC (2016) Phylogenomics reveals three sources of adaptive variation during a rapid irradiation. PLoS Biol 14(2):e1002379
Pellegrino KC, Rodrigues MT, Waite AN, Morando M, Yassuda YY, Sites JW Jr (2005) Phylogeography and species limits in the Gymnodactylus darwinii complex (Gekkonidae, Squamata) genetic structure coincides with river systems in the Brazilian Atlantic Forest. Biol J Linn Soc 85(1):13–26
Pirani RM, Tonini JF, Thomaz AT, Napoli MF, Encarnação LC, Knowles LL, Werneck FP (2022) Deep genomic divergence and phenotypic admixture of the treefrog Dendropsophus elegans (Hylidae: Amphibia) coincide with riverine boundaries at the Brazilian Atlantic Forest. Front Ecol Evol 10:765977
Rastogi A, Stróżecki M, Kalaji HM, Łuców D, Lamentowicz M, Juszczak R (2019) Impact of warming and reduced precipitation on photosynthetic and remote sensing properties of peatland vegetation. Environ Exp Bot 160:71–80
Revell LJ (2012) phytools an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3(2):217–223
Ribeiro MC, Metzger JP, Martensen AC, Ponzoni FJ, Hirota MM (2009) The Brazilian Atlantic Forest how much is left, and how is the remaining forest distributed? Implications for conservation. Biol Conserv 142(6):1141–1153
Ringelberg JJ, Koenen EJ, Sauter B, Aebli A, Rando JG, Iganci JR et al (2023) Precipitation is the main axis of tropical plant phylogenetic turnover across space and time. Sci Adv 9(7):eade4954
Schneider K, Adams CE, Elmer KR (2019) Parallel selection on ecologically relevant gene functions in the transcriptomes of highly diversifying salmonids. BMC Genom 20(1):1010
Schröder F, Lisso J, Müssig C (2011) EXORDIUM-LIKE1 promotes growth during low carbon availability in Arabidopsis. Plant Physiol 156(3):1620–1630
Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212
Smith SD, Pennell MW, Dunn CW, Edwards SV (2020) Phylogenetics is the new genetics (for most of biodiversity). Trends Ecol Evol 35(5):415–425
Stam R, Silva-Arias GA, Tellier A (2019) Subsets of NLR genes show differential signatures of adaptation during colonization of new habitats. New Phytol 224(1):367–379
Strelin MM, Zattara EE, Ullrich K, Schallenberg-Rüdinger M, Rensing S (2022) Delayed differentiation of epidermal cells walls can underlie pedomorphosis in plants the case of pedomorphic petals in the hummingbird-pollinated Caiophora hibiscifolia (Loasaceae, subfam Loasoideae) species. EvoDevo 13:1–14
Suetsugu K, Fukushima K, Makino T, Ikematsu S, Sakamoto T, Kimura S (2022) Transcriptomic heterochrony and completely cleistogamous flower development in the mycoheterotrophic orchid Gastrodia. New Phytol 237(1):323–338
Taylor NP, Zappi DC (2004) Cacti of Eastern Brazil, 1st edn. Royal Botanic Gardens, Kew, p 499
Taylor NP, Zappi DC, Romeiro-Brito M, Telhe MC, Franco FF, Moraes EM (2023) A phylogeny of Cereus (Cactaceae) and the placement and description of two new species. Taxon 72(6):1321–1333
Thomé MTC, Zamudio KR, Giovanelli JG, Haddad CF, Baldissera FA Jr, Alexandrino J (2010) Phylogeography of endemic toads and post-Pliocene persistence of the Brazilian Atlantic Forest. Mol Phylogenet Evol 55(3):1018–1031
Timpte C (2001) Auxin binding protein curiouser and curiouser. Trends Plant Sci 6(12):586–590
Vandenbrink JP, Kiss JZ, Herranz R, Medina FJ (2014) Light and gravity signals synergize in modulating plant development. Front Plant Sci 5:563
Warakanont J, Tsai CH, Michel EJ, Murphy GR III, Hsueh PY, Roston RL et al (2015) Chloroplast lipid transfer processes in Chlamydomonas reinhardtii involving a trigalactosyldiacykgkycerol 2 (TGD 2) orthologue. Plant J 84(5):1005–1020
Weigel RR, Bäuscher C, Pfitzner AJ, Pfitzner UM (2001) NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol Biol 46:143–160
Westerband AC, Funk JL, Barton KE (2021) Intraspecific trait variation in plants a renewed focus on its role in ecological processes. Ann Bot 127(4):397–410
Wu M, Kostyun JL, Hahn MW, Moyle LC (2018) Dissecting the basis of novel trait evolution in a radiation with widespread phylogenetic discordance. Mol Ecol 27(16):3301–3316
Yang Z (2007) PAML 4 phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang Z, Nielsen R (2002) Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol 19(6):908–917
Yang L, Zhao Y, Zhang Q, Cheng L, Han M, Ren Y, Yang L (2019) Effects of drought–re-watering–drought on the photosynthesis physiology and secondary metabolite production of Bupleurum chinense DC. Plant Cell Rep 38(9):1181–1197
Zamudio KR, Bell RC, Mason NA (2016) Phenotypes in phylogeography: species’ traits, environmental variation, and vertebrate diversification. Proc Natl Acad Sci USA 113(29):8041–8048
Zhang H, Chen J, Zhang F, Song Y (2019) Transcriptome analysis of callus from melon. Gene 684:131–138
Zhao XX, Huang LK, Zhang XQ, Li Z, Peng Y (2014) Effects of heat acclimation on photosynthesis, antioxidant enzyme activities, and gene expression in orchardgrass under heat stress. Molecules 19(9):13564–13576
Zhu W, Gao E, Shaban M, Wang Y, Wang H, Nie X, Zhu L (2018) GhUMC1, a blue copper-binding protein, regulates lignin synthesis and cotton immune response. Biochem Biophys Res Commun 504(1):75–81
Zizka A, Carvalho-Sobrinho JG, Pennington RT, Queiroz LP, Alcantara S, Baum DA et al (2020) Transitions between biomes are common and directional in Bombacoideae (Malvaceae). J Biogeogr 47(6):1310–1321
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data [online]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Berwal M, Ram C (2018) Superoxide dismutase: a stable biochemical marker for abiotic stress tolerance in higher plants. In: de Oliveira AB (ed) Abiotic and biotic stress in plants. IntechOpen. https://doi.org/10.5772/intechopen.82079
Demain AL, Fang A (2000) The natural functions of secondary metabolites. In: Fiechter A (ed) History of modern biotechnology I. Advances in biochemical engineering/biotechnology, vol 69, pp 1–39
Hannon GJ (2010) FASTX-Toolkit. http://hannonlab.cshl.edu/fastx_toolkit
Acknowledgements
We acknowledge Gerardus Olsthoorn for his support in the sampling of plant material. We acknowledge the support of São Paulo Research Foundation (FAPESP; 2018/03428-5 to F.F.F and E.M.M; 2020/15161-3 to F.F.F; and 2022/09910-9 to D.T.A.) and National Council for Scientific and Technological Development (CNPq) (402209/2016-8 to F.F.F., 310962/2022-6 to E.M.M. and 304178/2021-7 to D.C.Z.). We also thank the fellowships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES, Finance Code 001) to D.T.A, and FAPESP to M.R.B and M.C.T (2018/06937-8 and 2019/11233-2). We also extend our gratitude to the Fernando de Noronha Marine National Park (ICMBIO/PARNAMAR) and the government administration of Fernando de Noronha (DEFN) for their assistance in sampling Cereus insularis.
Author information
Authors and Affiliations
Contributions
The idea for this study was conceived by FFF and DTA. Data collection and analyses were performed by DTA, MRB, and MCT. DTA led the writing of the paper, while FFF, IASB, EMM, NT, and DZ contributed with numerous conceptions and writing. All the authors contributed to the intellectual development of the paper, made multiple revisions, and approved the final draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Amaral, D.T., Bonatelli, I.A.S., Romeiro-Brito, M. et al. Comparative transcriptome analysis reveals lineage- and environment-specific adaptations in cacti from the Brazilian Atlantic Forest. Planta 260, 4 (2024). https://doi.org/10.1007/s00425-024-04442-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-024-04442-x