Abstract
Main conclusion
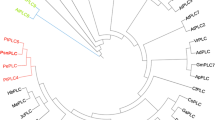
A phospholipase Dα gene ( AnPLDα ) was cloned from xerophytic desert plant Ammopiptanthus nanus and its overexpression enhanced salt tolerance of a PLDα1 deficient Arabidopsis mutant.
Phospholipase Dα (PLDα) hydrolyzes phosphatidylcholine to produce phosphatidic acid, and plays crucial role in plant tolerance to abiotic stress. In this study, a phospholipase Dα gene (AnPLDα) was cloned from xerophyte Ammopiptanthus nanus by the methods of homologous cloning and rapid amplification of cDNA ends, and evaluated for its function in stress tolerance. The full-length cDNA was 2832 bp long, containing an open reading frame of 2427 bp that encodes 808 amino acids. The putative protein was predicted to be localized to the cytoplasm and this was confirmed by transient expression of a fluorescent fusion protein. The endogenous expression of the AnPLDα gene was induced by high salt, dehydration, cold and abscisic acid. The heterologous expression of the AnPLDα gene improved salt tolerance of an Arabidopsis pldα1 knocked out mutant, and positively regulated the expression of the AtABI, AtNCED, AtRD29A, AtRD29B and AtADH genes. Therefore, the AnPLDα gene was concluded to be involved in response to abiotic stress. The AnPLDα gene is a hopeful candidate for transgenic application to improve stress tolerance of commercial crops.











Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- eGFP:
-
Enhanced green fluorescent protein
- ORF:
-
Open reading frame
- PA:
-
Phosphatidic acid
- PLDα:
-
Phospholipase Dα
References
Bargmann BOR, Laxalt AM, Riet BT, Schooten BV, Merquiol E, Testerink C, Haring MA, Bartels D, Munnik T (2009) Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol 50:78–89
Barrero JM, Rodriguez PL, Quesada V, Piqueras P, Ponce MR, Micol JL (2006) Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ 29:2000–2008
Chen GQ, Huang HW, Kang M, Ge XJ (2007) Development and characterization of microsatellite markers for an endangered shrub, Ammopiptanthus mongolicus (Leguminosae) and cross-species amplification in Ammopiptanthus nanus. Conserv Genet 8:1495–1497
Chen X, Wang YF, Lv B, Li J, Luo LQ, Lu SC, Zhang X, Ma H, Ming F (2014) The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol 55:604–619
Cheng SH (1959) Ammopiptanthus Cheng f. a new genus of Leguminosae from central Asia. J Bot USSR 44:1381–1386
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Deng LQ, Yu HQ, Liu YP, Jiao PP, Zhou SF, Zhang SZ, Li WC, Fu FL (2014) Heterologous expression of antifreeze protein gene AnAFP from Ammopiptanthus nanus enhances cold tolerance in Escherichia coli and tobacco. Gene 539:132–140
Devaiah SP, Pan X, Hong Y, Roth M, Welti R, Wang XM (2007) Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis. Plant J 50:950–957
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull 19:11–15
Eliáš M, Potocký M, CvrčkováF Žárský V (2002) Molecular diversity of phospholipase D in angiosperms. BMC Genom 3:2
Fan L, Zheng SQ, Wang XM (1997) Antisense suppression of phospholipase Dα retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9:2183–2196
Garnier J, Gibrat JF, Robson B (1996) GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol 266:540–553
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook. Human Press, New York, pp 571–607
Ge XJ, Yu Y, Yuan YM, Huang HW, Yan C (2005) Genetic diversity and geographic differentiation in endangered Ammopiptanthus populations in desert regions of northwest China as revealed by ISSR analysis. Ann Bot 95:843–851
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30:1229–1235
Hong YY, Pan XQ, Welti R, Wang XM (2008a) Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20:803–816
Hong YY, Zheng SQ, Wang XM (2008b) Dual functions of phospholipase Dα1 in plant response to drought. Mol Plant 1:262–269
Hsieh TH, Li CW, Su RC, Cheng CP, Sanjaya Tsai YC, Chan MT (2010) A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 231:1459–1473
Huang YY, Shi Y, Lei Y, Li Y, Fan J, Xu YJ, Ma XF, Zhao JQ, **ao SY, Wang WM (2013) Functional identification of multiple nucleocytoplasmic trafficking signals in the broad spectrum resistance protein RPW8.2. Planta 239:455–468
Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 24:1695–1708
Jiang YJ, Liang G, Yu DQ (2012) Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol Plant 5:1375–1388
Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, Todaka D, Nakashima K, Hirayama T, Shinozaki K, Yamaguchi-Shinozaki K (2011) An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol 52:2136–2146
Kolesnikov YS, Nokhrina KP, Kretynin SV, Volotovski ID, Martinec J, Romanov GA, Kravets VS (2012) Molecular structure of phospholipase D and regulatory mechanisms of its activity in plant and animal cells. Biochemistry (Mosc) 77:1–14
Kooijman EE, Chupin V, de Kruijff B, Burger KN (2003) Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 4:162–174
Lee SJ, Kang JY, Park HJ, Kim MD, Bae MS, Choi HI, Kim SY (2010) DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol 153:716–727
Lee JH, Welti R, Roth M, Schapaugh WT, Li JR, Trick HN (2012) Enhanced seed viability and lipid compositional changes during natural aging by suppressing phospholipase Dα in soybean seed. Plant Biotechnol J 10:164–173
Li G, Lin F, Xue HW (2007) Genome-wide analysis of the phospholipase D family in Oryza sativa and functional characterization of PLDβ1 in seed germination. Cell Res 17:881–894
Li MY, Hong YY, Wang XM (2009) Phospholipase D- and phosphatidic acid- mediated signaling in plants. Biochim Biophys Acta 1791:927–935
Li JB, Luan YS, Liu Z (2014) Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol Plant. doi:10.1111/ppl.12315
Liu B, Yao L, Wang WG, Gao JH, Chen F, Wang SH, Xu Y, Tang L, Jia YJ (2010) Molecular cloning and characterization of phospholipase D from Jatropha curcas. Mol Biol Rep 37:939–946
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324:1064–1068
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:222–226
Min JH, Chung JS, Lee KH, Kim CS (2015) The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J Integr Plant Biol 57:313–324
Peng YL, Zhang JP, Cao GY, **e YH, Liu XH, Lu MH, Wang GY (2010) Overexpression of a PLDα1 gene from Setaria italica enhances the sensitivity of Arabidopsis to abscisic acid and improves its drought tolerance. Plant Cell Rep 29:793–802
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Qiu HL, Zhang LH, Liu C, He L, Wang AY, Liu HL, Zhu JB (2014) Cloning and characterization of a novel dehydrin gene, SiDhn2, from Saussurea involucrata Kar. et Kir. Plant Mol Biol 84:707–718
Ruelland E, Kravets V, Derevyanchuk M, Martinec J, Zachowski A, Pokotylo I (2015) Role of phospholipid signalling in plant environmental responses. Environ Exp Bot 114:129–143
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2000) Overexpression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Saijo Y, Kinoshita N, Ishiyama K, Hata S, Kyozuka J, Hayakawa T, Nakamura T, Shimamoto K, Yamaya T, Izui K (2001) A Ca2+-dependent protein kinase that endows rice plants with cold- and salt-stress tolerance functions in vascular bundles. Plant Cell Physiol 42:1228–1233
Sharma P, Jha AB, Dubey RS, Pessarakli M (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Bot, ID 217037
Shen P, Wang R, **g W, Zhang WH (2011) Rice phospholipase Dα is involved in salt tolerance by the mediation of H+-ATPase activity and transcription. J Integr Plant Biol 53:289–299
Soon FF, Ng LM, Zhou XE, West GM, Kovach A, Tan MH, Suino-Powell KM, He Y, Xu Y, Chalmers M, Brunzelle JS, Zhang H, Yang H, Jiang H, Li J, Yong EL, Cutler S, Zhu JK, Griffin PR, Melcher K, Xu HE (2012) Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335:85–88
Szczegielniak J, Klimecka M, Liwosz A, Ciesielski A, Kaczanowski S, Dobrowolska G, Harmon AC, Muszynska G (2005) A wound responsive and phospholipid-regulated maize calcium-dependent protein kinase. Plant Physiol 139:1970–1983
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tuteja N (2007) Mechanisms of high salinity tolerance in plants. Methods Enzymol 428:419–438
Wang XM (2000) Multiple forms of phospholipase D in plants: the gene family, catalytic and regulatory properties, and cellular functions. Prog Lipid Res 39:109–149
Wang XM (2001) Plant phospholipases. Annu Rev Plant Biol 52:211–231
Wang ZJ (2005) Survey and protection for rare plant resource Ammopiptanthus nanus. Chin Wild Plant Resour 24:41–42
Wang XM, Devaiah SP, Zhang WH, Welti R (2006) Signaling functions of phosphatidic acid. Prog Lipid Res 45:250–278
Wang W, Chen JJ, Li JN, Zhang YH, Shao ZY, Kuai BK (2007) Extraordinary accumulations of antioxidants in Ammopiptanthus mongolicus (Leguminosae) and Tetraena mongolica (Zygophyllaceae) distributed in extremely stressful environments. Bot Stud 48:55–61
Wang JB, Ding B, Guo YL, Li M, Chen SJ, Huang GZ, **e XD (2014) Overexpression of a wheat phospholipase D gene, TaPLDα, enhances tolerance to drought and osmotic stress in Arabidopsis thaliana. Planta 240:103–115
Xu ZY, Kim SY, de Hyeon Y, Kim DH, Dong T, Park Y, ** JB, Joo SH, Kim SK, Hong JC, Hwang D, Hwang I (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25:4708–4724
Yang JD, Worley E, Udvardi M (2014) A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26:4862–4874
Yang N, Ding FX, Wu GF, Wang CL, Ding L, An LZ (2015) Phospholipase Dα from Chorispora bungeana: cloning and partial functional characterization. Plant Growth Regul 75:511–520
Ying S, Zhang DF, Fu J, Shi YS, Song YC, Wang TY, Li Y (2012) Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 235:253–266
Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Four Arabidopsis/AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant, Cell Environ 38:35–49
You XR, Zhang YY, Li L, Li ZC, Li MJ, Li CB, Zhu JH, Peng HX, Sun J (2014) Cloning and molecular characterization of phospholipase D (PLD)delta gene from longan (Dimocarpus longan Lour.). Mol Biol Rep 41:4351–4360
Yu LJ, Nie JN, Cao CY, ** YK, Yan M, Wang FZ, Liu J, **ao Y, Liang YH, Zhang WH (2010) Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol 188:762–773
Yu HQ, Wang YG, Yong TM, She YH, Fu FL, Li WC (2014) Heterologous expression of betaine aldehyde dehydrogenase gene from Ammopiptanthus nanus confers high salt and heat tolerance to Escherichia coli. Gene 549:77–84
Yu HQ, Zhang YY, Yong TM, Liu YP, Zhou SF, Fu FL, Li WC (2015a) Cloning and functional validation of molybdenum cofactor sulfurase gene from Ammopiptanthus nanus. Plant Cell Rep 34:1165–1176
Yu XM, Li AH, Li WQ (2015b) How membranes organize during seed germination: three patterns of dynamic lipid remodelling define chilling resistance and affect plastid biogenesis. Plant Cell Environ 38:1391–1403
Zhang WH, Qin CB, Zhao J, Wang XM (2004) Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101:9508–9513
Zhang Q, Lin F, Mao TL, Nie JN, Yan M, Yuan M, Zhang WH (2012) Phosphatidic acid regulates microtubule organization by interacting with map65-1 in response to salt stress in Arabidopsis. Plant Cell 24:4555–4576
Zhao J (2015) Phospholipase D and phosphatidic acid in plant defence response: from protein-protein and lipid-protein interactions to hormone signaling. J Exp Bot 66:1721–1736
Zhao JZ, Zhou D, Zhang Q, Zhang WH (2012) Genomic analysis of phospholipase D family and characterization of GmPLDαs in soybean (Glycine max). J Plant Res 125:569–578
Zheng L, Krishnamoorthi R, Zolkiewski M, Wang XM (2000) Distinct Ca2+ binding properties of novel C2 domains of plant phospholipase D. J Biol Chem 275:19700–19706
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgments
This work was supported by the National Key Science and Technology Special Project (2014ZX08003-004) and the National Natural Science Foundation of China (31071433). The authors thank the technical support from the Key Laboratory of Biology and Genetic Improvement of Maize in Southwest Region.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
H. Q. Yu and T. M. Yong contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, H.Q., Yong, T.M., Li, H.J. et al. Overexpression of a phospholipase Dα gene from Ammopiptanthus nanus enhances salt tolerance of phospholipase Dα1-deficient Arabidopsis mutant. Planta 242, 1495–1509 (2015). https://doi.org/10.1007/s00425-015-2390-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2390-5