Abstract
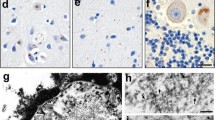
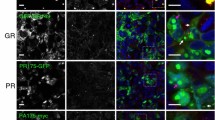
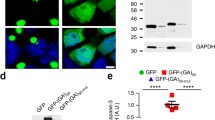
Neuronal intranuclear inclusion disease (NIID) is neurodegenerative disease characterized by widespread inclusions. Despite the identification of GGC repeat expansion in 5’UTR of NOTCH2NLC gene in adult-onset NIIDs, its pathogenic mechanism remains unclear. Gain-of-function poly-amino-acid proteins generated by unconventional translation have been revealed in nucleotide repeat expansion disorders, inspiring us to explore the possibility of unconventional translation in NIID. Here we demonstrated that NOTCH2NLC 5’UTR triggers the translation of a polyglycine (polyG)-containing protein, N2NLCpolyG. N2NLCpolyG accumulates in p62-positive inclusions in cultured cells, mouse models, and NIID patient tissues with NOTCH2NLC GGC expansion. Translation of N2NLCpolyG is initiated by an upstream open reading frame (uORF) embedding the GGC repeats. N2NLCpolyG tends to aggregate with the increase of GGC repeat units, and displays phase separation properties. N2NLCpolyG aggregation impairs nuclear lamina and nucleocytoplasmic transport but does not necessarily cause acute death on neuronal cells. Our study suggests a similarity of pathogenic mechanisms between NIID and another GGC-repeat disease, fragile X-associated tremor ataxia syndrome. These findings expand our knowledge of protein gain-of-function in NIID, and further highlight evidence for a novel spectrum of diseases caused by aberrant polyG protein aggregation, namely the polyG diseases.







Similar content being viewed by others
Abbreviations
- 5’UTR:
-
5’-Untranslated regions
- ADLD:
-
Adult-onset autosomal-dominant leukodystrophy
- CDS:
-
Coding sequence
- FXTAS:
-
Fragile X-associated tremor/ataxia syndrome
- GFP:
-
Green fluorescence protein
- IUE:
-
In-utero electroporation
- NIID:
-
Neuronal intranuclear inclusion disease
- LLPS:
-
Liquid-to-liquid phase separation
- NREDs:
-
Nucleotide repeat expansion disorders
- polyA RNA:
-
Polyadenylated RNA
- polyG:
-
Polyglycine
- RAN:
-
Repeat-associated non-AUG
- ribo-seq:
-
Ribosome profiling
- uORF:
-
Upstream open reading frame
References
Ambadipudi S, Biernat J, Riedel D, Mandelkow E, Zweckstetter M (2017) Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat Commun 8:275. https://doi.org/10.1038/s41467-017-00480-0
Asamitsu S, Yabuki Y, Ikenoshita S, Kawakubo K, Kawasaki M, Usuki S et al (2021) CGG repeat RNA G-quadruplexes interact with FMRpolyG to cause neuronal dysfunction in fragile X-related tremor/ataxia syndrome. Sci Adv. https://doi.org/10.1126/sciadv.abd9440
Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646. https://doi.org/10.1016/j.neuron.2013.02.004
Boivin M, Deng J, Pfister V, Grandgirard E, Oulad-Abdelghani M, Morlet B et al (2021) Translation of GGC repeat expansions into a toxic polyglycine protein in NIID defines a novel class of human genetic disorders: the polyG diseases. Neuron 109:1825-1835.e1825. https://doi.org/10.1016/j.neuron.2021.03.038
Boivin M, Pfister V, Gaucherot A, Ruffenach F, Negroni L, Sellier C et al (2020) Reduced autophagy upon C9ORF72 loss synergizes with dipeptide repeat protein toxicity in G4C2 repeat expansion disorders. EMBO J 39:e100574. https://doi.org/10.15252/embj.2018100574
Chen Z, Yan Yau W, Jaunmuktane Z, Tucci A, Sivakumar P, Gagliano Taliun SA et al (2020) Neuronal intranuclear inclusion disease is genetically heterogeneous. Ann Clin Transl Neurol 7:1716–1725. https://doi.org/10.1002/acn3.51151
Chou CC, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, Liu F et al (2018) TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat Neurosci 21:228–239. https://doi.org/10.1038/s41593-017-0047-3
D’Angelo MA, Raices M, Panowski SH, Hetzer MW (2009) Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136:284–295. https://doi.org/10.1016/j.cell.2008.11.037
Deng J, Gu M, Miao Y, Yao S, Zhu M, Fang P et al (2019) Long-read sequencing identified repeat expansions in the 5’UTR of the NOTCH2NLC gene from Chinese patients with neuronal intranuclear inclusion disease. J Med Genet 56:758–764. https://doi.org/10.1136/jmedgenet-2019-106268
Deng J, Yu J, Li P, Luan X, Cao L, Zhao J et al (2020) Expansion of GGC repeat in GIPC1 Is associated with oculopharyngodistal myopathy. Am J Hum Genet 106:793–804. https://doi.org/10.1016/j.ajhg.2020.04.011
Fiddes IT, Lodewijk GA, Mooring M, Bosworth CM, Ewing AD, Mantalas GL et al (2018) Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173:1356-1369.e1322. https://doi.org/10.1016/j.cell.2018.03.051
Finnsson J, Sundblom J, Dahl N, Melberg A, Raininko R (2015) LMNB1-related autosomal-dominant leukodystrophy: clinical and radiological course. Ann Neurol 78:412–425. https://doi.org/10.1002/ana.24452
Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH et al (2015) GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525:129–133. https://doi.org/10.1038/nature14974
Fukai Y, Yorimitsu D, Nishimura H, Kutoku Y, Sasaki T, Kashihara N et al (2018) Proteinuria in neuronal intranuclear inclusion disease. Neurol Clin Neurosci. https://doi.org/10.1111/ncn3.12182
Gao FB, Richter JD, Cleveland DW (2017) Rethinking unconventional translation in neurodegeneration. Cell 171:994–1000. https://doi.org/10.1016/j.cell.2017.10.042
Gelpi E, Botta-Orfila T, Bodi L, Marti S, Kovacs G, Grau-Rivera O et al (2017) Neuronal intranuclear (hyaline) inclusion disease and fragile X-associated tremor/ataxia syndrome: a morphological and molecular dilemma. Brain: J Neurol 140:e51. https://doi.org/10.1093/brain/awx156
Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ et al (2017) RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat Commun 8:2005. https://doi.org/10.1038/s41467-017-02200-0
Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J (2001) Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology 57:127–130. https://doi.org/10.1212/wnl.57.1.127
Hinnebusch AG, Ivanov IP, Sonenberg N (2016) Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science (New York, NY) 352:1413–1416. https://doi.org/10.1126/science.aad9868
Huang S, Zhu S, Li XJ, Li S (2019) The expanding clinical universe of polyglutamine disease. Neurosci: Rev J Bringing Neurobiolneurol Psychiatry 25:512–520. https://doi.org/10.1177/1073858418822993
Hukema RK, Buijsen RA, Raske C, Severijnen LA, Nieuwenhuizen-Bakker I, Minneboo M et al (2014) Induced expression of expanded CGG RNA causes mitochondrial dysfunction in vivo. Cell Cycle (Georgetown, Tex) 13:2600–2608. https://doi.org/10.4161/15384101.2014.943112
Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS (2012) The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7:1534–1550. https://doi.org/10.1038/nprot.2012.086
Ishiura H, Shibata S, Yoshimura J, Suzuki Y, Qu W, Doi K et al (2019) Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlap** disease. Nat Genet 51:1222–1232. https://doi.org/10.1038/s41588-019-0458-z
** P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC et al (2007) Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron 55:556–564. https://doi.org/10.1016/j.neuron.2007.07.020
** P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K et al (2003) RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron 39:739–747. https://doi.org/10.1016/s0896-6273(03)00533-6
Jovičić A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB et al (2015) Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18:1226–1229. https://doi.org/10.1038/nn.4085
Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC et al (2016) CGG repeat-associated non-AUG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol Cell 62:314–322. https://doi.org/10.1016/j.molcel.2016.02.034
Krishnan G, Zhang Y, Gu Y, Kankel MW, Gao FB, Almeida S (2020) CRISPR deletion of the C9ORF72 promoter in ALS/FTD patient motor neurons abolishes production of dipeptide repeat proteins and rescues neurodegeneration. Acta Neuropathol 140:81–84. https://doi.org/10.1007/s00401-020-02154-6
Li N, Lagier-Tourenne C (2018) Nuclear pores: the gate to neurodegeneration. Nat Neurosci 21:156–158. https://doi.org/10.1038/s41593-017-0066-0
Lin ST, Fu YH (2009) miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis Model Mech 2:178–188. https://doi.org/10.1242/dmm.001065
Lu X, Hong D (2021) Neuronal intranuclear inclusion disease: recognition and update. J Neural Transm (Vienna, Austria 1996) 128:295–303. https://doi.org/10.1007/s00702-021-02313-3
Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, Needham PG et al (2019) RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102:321-338.e328. https://doi.org/10.1016/j.neuron.2019.01.048
Michel AM, Fox G, A MK, De Bo C, O’Connor PB, Heaphy SM et al (2014) GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res 42:D859-864. https://doi.org/10.1093/nar/gkt1035
Michel AM, Kiniry SJ, O’Connor PBF, Mullan JP, Baranov PV (2018) GWIPS-viz: 2018 update. Nucleic Acids Res 46:D823-d830. https://doi.org/10.1093/nar/gkx790
Minakawa EN, Popiel HA, Tada M, Takahashi T, Yamane H, Saitoh Y et al (2020) Arginine is a disease modifier for polyQ disease models that stabilizes polyQ protein conformation. Brain: J Neurol 143:1811–1825. https://doi.org/10.1093/brain/awaa115
Motoki M, Nakajima H, Sato T, Tada M, Kakita A, Arawaka S (2018) Neuronal intranuclear inclusion disease showing intranuclear inclusions in renal biopsy 12 years earlier. Neurology 91:884–886. https://doi.org/10.1212/wnl.0000000000006480
Ogasawara M, Iida A, Kumutpongpanich T, Ozaki A, Oya Y, Konishi H et al (2020) CGG expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy with neurological manifestations. Acta Neuropathol Commun 8:204. https://doi.org/10.1186/s40478-020-01084-4
Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY et al (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162:1066–1077. https://doi.org/10.1016/j.cell.2015.07.047
Rodriguez CM, Todd PK (2019) New pathologic mechanisms in nucleotide repeat expansion disorders. Neurobiol Dis 130:104515. https://doi.org/10.1016/j.nbd.2019.104515
Saito T (2006) In vivo electroporation in the embryonic mouse central nervous system. Nat Protoc 1:1552–1558. https://doi.org/10.1038/nprot.2006.276
Sellier C, Buijsen RAM, He F, Natla S, Jung L, Tropel P et al (2017) Translation of expanded CGG repeats into FMRpolyG Is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron 93:331–347. https://doi.org/10.1016/j.neuron.2016.12.016
Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F et al (2013) Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep 3:869–880. https://doi.org/10.1016/j.celrep.2013.02.004
Sone J, Mitsuhashi S, Fujita A, Mizuguchi T, Hamanaka K, Mori K et al (2019) Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat Genet 51:1215–1221. https://doi.org/10.1038/s41588-019-0459-y
Sone J, Mori K, Inagaki T, Katsumata R, Takagi S, Yokoi S et al (2016) Clinicopathological features of adult-onset neuronal intranuclear inclusion disease. Brain: J Neurol 139:3170–3186. https://doi.org/10.1093/brain/aww249
Sugiyama A, Sato N, Kimura Y, Maekawa T, Enokizono M, Saito Y (2017) MR imaging features of the cerebellum in adult-onset neuronal intranuclear inclusion disease: 8 cases. AJNR Am J Neuroradiol 38:2100–2104. https://doi.org/10.3174/ajnr.A5336
Sun QY, Xu Q, Tian Y, Hu ZM, Qin LX, Yang JX et al (2020) Expansion of GGC repeat in the human-specific NOTCH2NLC gene is associated with essential tremor. Brain: J Neurol 143:222–233. https://doi.org/10.1093/brain/awz372
Suzuki IK, Gacquer D, Van Heurck R, Kumar D, Wojno M, Bilheu A et al (2018) Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell 173:1370-1384.e1316. https://doi.org/10.1016/j.cell.2018.03.067
Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee CZ et al (2018) CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat Commun 9:152. https://doi.org/10.1038/s41467-017-02643-5
Tian Y, Wang JL, Huang W, Zeng S, Jiao B, Liu Z et al (2019) Expansion of human-specific GGC repeat in neuronal intranuclear inclusion disease-related disorders. Am J Hum Genet 105:166–176. https://doi.org/10.1016/j.ajhg.2019.05.013
Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M et al (2013) CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78:440–455. https://doi.org/10.1016/j.neuron.2013.03.026
Wei S, Du H, Li Z, Tao G, Xu Z, Song X et al (2019) Transcription factors Sp8 and Sp9 regulate the development of caudal ganglionic eminence-derived cortical interneurons. J Comp Neurol 527:2860–2874. https://doi.org/10.1002/cne.24712
Westenberger A, Klein C (2020) Essential phenotypes of NOTCH2NLC-related repeat expansion disorder. Brain: J Neurol 143:5–8. https://doi.org/10.1093/brain/awz404
** J, Wang X, Yue D, Dou T, Wu Q et al (2021) 5’ UTR CGG repeat expansion in GIPC1 is associated with oculopharyngodistal myopathy. Brain: J Neurol 144:601–614. https://doi.org/10.1093/brain/awaa426
Yu J, Deng J, Guo X, Shan J, Luan X, Cao L et al (2021) The GGC repeat expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy type 3. Brain: J Neurol. https://doi.org/10.1093/brain/awab077
Yu J, Deng J, Guo X, Shan J, Luan X, Cao L, Zhao J, Yu M, Zhang W, Lv H et al (2021) The GGC repeat expansion in NOTCH2NLC is associated with oculopharyngodistal myopathy type 3. Brain: J Neurol 144:1819–1832. https://doi.org/10.1093/brain/awab077
Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L et al (2014) Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol 128:505–524. https://doi.org/10.1007/s00401-014-1336-5
Zhong S, Zhao Z, **e W, Cai Y, Zhang Y, Ding J, Wang X (2021) GABAergic interneuron and neurotransmission are mTOR-dependently disturbed in experimental focal cortical dysplasia. Mol Neurobiol 58:156–169. https://doi.org/10.1007/s12035-020-02086-y
Acknowledgements
This work was supported by project grants from the National Natural Science Foundation of China (Code: 81771308, 31771184). We thank the patients and control subjects for participating in the study. We also thank the neurologists, pathologists and radiologists for their help in collecting clinical information, especially Dr. Yang Cai, Dr. ** Zhong, Yangye Lian, Wenyi Luo, **-Zhong-Aff1">