Abstract
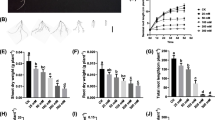
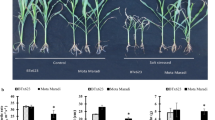
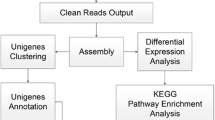
Soil salinity influences various aspects of crop growth and development. Identification of novel germplasm and genetic regulators of salt tolerance will help to alleviate the threat of land salinization. Here, we identified salt-tolerant SHEN5003 and -sensitive YZ26 maize germplasm from different heterotic groups. Salt-grown SHEN5003 (PA group) lines exhibited decreased reduction in fresh weight and dry weight, high SPAD value, total sugar and K+ contents, as well as low Na+ content relative to salt-treated YZ26 (BSSS group) plants. Salt-tolerant SHEN5003 and -sensitive YZ26 lines were followingly used for the discovery of genetic regulators of salt tolerance. Transcriptome outcome showed the expression of genes relevant to Na+/K+ homeostasis, such as H+-ATPase, HKT1, and HAKs together with CAT2 for ROS detoxification was altered in salt-treated SHEN5003 line. Moreover, GO enrichment analysis indicated differentially expressed genes of SHEN5003 upon salt treatment was significantly enriched in the stress response pathway (GO:0006950). By integrated transcriptome analyses of SHEN5003 and YZ26 lines grown in normal and salinity conditions, positive regulator of salt tolerance ZmSWEET1b was revealed, which exhibited the opposite expression pattern between salt-treated SHEN5003 and YZ26 lines. ZmSWEET1b interacted with multiple partners, such as Sucrose synthase 2 to shape a functional association network in the modulation of sugar metabolism upon salinity stress.







Similar content being viewed by others
References
Bazihizina N, Colmer TD, Cuin TA, Mancuso S, Shabala S (2019) Friend or foe? Chloride patterning in halophytes. Trends Plant Sci 24:142–151
Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123:1279–1291
Cao Y, Liang X, Yin P, Zhang M, Jiang C (2019) A domestication-associated reduction in K+-preferring HKT transporter activity underlies maize shoot K+ accumulation and salt tolerance. New Phytol 222:301–317
Cheng X, Zhang S, Tao W, Zhang X, Liu J, Sun J, Zhang H, Pu L, Huang R, Chen T (2018) INDETERMINATE SPIKELET1 recruits histone deacetylase and a transcriptional repression complex to regulate rice salt tolerance. Plant Physiol 178:824–837
Chow PS, Landhäusser SM (2004) A method for routine measurements of total sugar and starch content in woody plant tissues. Tree Physiol 24:1129–1136
Demidchik V, Maathuis FJ (2007) Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytol 175:387–404
Eggert E, Obata T, Gerstenberger A, Gier K, Brandt T, Fernie AR, Schulze W, Kühn C (2016) A sucrose transporter-interacting protein disulphide isomerase affects redox homeostasis and links sucrose partitioning with abiotic stress tolerance. Plant Cell Environ 39:1366–1380
El Mahi H, Pérez-Hormaeche J, De Luca A, Villalta I, Espartero J, Gámez-Arjona F, Fernández JL, Bundó M, Mendoza I, Mieulet D, Lalanne E, Lee SY, Yun DJ, Guiderdoni E, Aguilar M, Leidi EO, Pardo JM, Quintero FJ (2019) A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol 180:1046–1065
Gong Z, **ong L, Shi H, Yang S, Herrera-Estrella LR, Xu G, Chao DY, Li J, Wang PY, Qin F, Li J, Ding Y, Shi Y, Wang Y, Yang Y, Guo Y, Zhu JK (2020) Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci 63:635–674
Ismail AM, Horie T (2017) Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu Rev Plant Biol 68:405–434
Jiang Z, Zhou X, Tao M, Yuan F, Liu L, Wu F, Wu X, **ang Y, Niu Y, Liu F, Li C, Ye R, Byeon B, Xue Y, Zhao H, Wang HN, Crawford BM, Johnson DM, Hu C, Pei C, Zhou W, Swift GB, Zhang H, Vo-Dinh T, Hu Z, Siedow JN, Pei ZM (2019) Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572:341–346
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Kumar S, Beena AS, Awana M, Singh A (2017) Salt-induced tissue-specific cytosine methylation downregulates expression of HKT genes in contrasting wheat (Triticum aestivum L.) genotypes. DNA Cell Biol 36:283–294
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25:402–408
Lu Y, Zhang S, Shah T, ** is a powerful approach to detecting quantitative trait loci underlying drought tolerance in maize. Proc Natl Acad Sci USA 107:19585–19590
Ma L, Ye J, Yang Y, Lin H, Yue L, Luo J, Long Y, Fu H, Liu X, Zhang Y, Wang Y, Chen L, Kudla J, Wang Y, Han S, Song CP, Guo Y (2019) The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev Cell 48:697–709
Meloni DA, Oliva MA, Martinez CA, Cambraia J (2003) Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ Exp Bot 49:69–76
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ 33:453–467
Monihan SM, Ryu CH, Magness CA, Schumaker KS (2019) Linking duplication of a calcium sensor to salt tolerance in Eutrema salsugineum. Plant Physiol 179:176–1192
Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37:1141–1146
Shabala S, Demidchik V, Shabala L, Cuin TA, Smith SJ, Miller AJ, Davies JM, Newman IA (2006) Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+-permeable channels. Plant Physiol 141:1653–1665
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
van Zelm E, Zhang Y, Testerink C (2020) Salt tolerance mechanisms of plants. Annu Rev Plant Biol 71:403–433
Wong TH, Li MW, Yao XQ, Lam HM (2013) The GmCLC1 protein from soybean functions as a chloride ion transporter. J Plant Physiol 170:101–104
Yamaguchi T, Blumwald E (2005) Develo** salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci 10:615–620
Yang Y, Guo Y (2018) Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol 217:523–539
Yang T, Zhang S, Hu Y, Wu F, Hu Q, Chen G, Cai J, Wu T, Moran N, Yu L, Xu G (2014) The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol 166:945–959
Yang Z, Wang C, Xue Y, Liu X, Chen S, Song C, Yang Y, Guo Y (2019) Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat Commun 10:1199
Yin YG, Kobayashi Y, Sanuki A, Kondo S, Fukuda N, Ezura H, Sugaya S, Matsukura C (2010) Salinity induces carbohydrate accumulation and sugar-regulated starch biosynthetic genes in tomato (Solanum lycopersicum L. cv. ’Micro-Tom’) fruits in an ABA- and osmotic stress-independent manner. J Exp Bot 61:563–574
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14
Zhang M, Cao Y, Wang Z, Wang ZQ, Shi J, Liang X, Song W, Chen Q, Lai J, Jiang C (2018) A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol 217:1161–1176
Zhang M, Liang X, Wang L, Cao Y, Song W, Shi J, Lai J, Jiang C (2019) A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat Plants 5:1297–1308
Zhang X, Liu P, Qing C, Yang C, Shen Y, Ma L (2021) Comparative transcriptome analyses of maize seedling root responses to salt stress. PeerJ 9:e10765
Zhang H, Zhu J, Gong Z, Zhu JK (2022) Abiotic stress responses in plants. Nat Rev Genet 23:104–119
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31571671), the High-end Talent Project of Yangzhou University (18HTYZU12), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Contributions
YJW: designed the research. SSW, WHD, YDC, and YWS: performed the experiments. YHD: analyzed the data. YJW and YHD: wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Handling Editor: Ranjit Riar.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
344_2023_11033_MOESM8_ESM.xlsx
Supplementary file8 (XLSX 13 KB) Differentially expressed genes with opposite expression patterns between YZ26 and SHEN5003 lines
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ding, Y., Wang, S., Du, W. et al. Identification of Germplasm and Sugar Transporter Gene ZmSWEET1b Associated with Salt Tolerance in Maize. J Plant Growth Regul 42, 7580–7590 (2023). https://doi.org/10.1007/s00344-023-11033-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-023-11033-9