Abstract
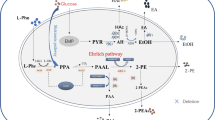
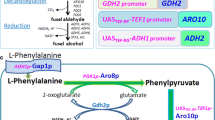
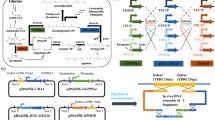
The 2-phenylethanol biosynthesis in Saccharomyces cerevisiae is limited by multiple-branch metabolism in the shikimate pathway. In this research, a total of 4 × 4 (sites × genes) guide sequences from four branch genes (TYR1, ARO8, AAT2 and ALD3) were designed. A single-gene down-regulation library of 4 × 4 Saccharomyces cerevisiae strains was constructed. By the assessment of gene expression level and 2-phenylethanol production, the optimal guide sequences of TYR1/AAT2/ALD3 were identified. On these bases, we first developed a high-yielding 2-phenylethanol strain carrying triple-CRISPRi system for simultaneous three branch repression. The INVScI-TYR1.AAT2.ALD3 successfully achieved the desired transcriptional repression effect and led to a 1.89-fold increase in 2-phenylethanol production compared to the starting strain. Triple-CRISPRi-mediated down-regulation of the shikimate pathway branch genes provided a convenient and efficient solution for the development of 2-phenylethanol high-yield Saccharomyces cerevisiae engineering strain.
Graphical abstract





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from corresponding author, [Huazhong Agricultural University], upon reasonable request.
References
Urszula D, Marcin O, Patrycja O et al (2015) Phase equilibrium and bioproduction of the aroma compound 2-phenylethanol in a biphasic aqueous system. Eur Food Res Technol. https://doi.org/10.1007/s00217-015-2421-2
Sara M, Mohamed K, Richard GM et al (2022) Bioproduction of 2-Phenylethanol through Yeast Fermentation on Synthetic Media and on Agro-Industrial Waste and By-Products: A Review. Foods. https://doi.org/10.3390/foods11010109
Wang CY, Yao YL, Li ES et al (2017) Synthesis of β - Phenylethanol. Flavour Fragrance Cosmetics 02:12–14
Wang, Zhuge ZB (2019) Advances in 2-phenylethanol production from engineered microorganisms. Biotechnology Advances: An International Review Journal. https://doi.org/10.1016/j.biotechadv.2019.02.005
Hua DL, Xu P (2011) Recent advances in biotechnological production of 2-phenylethanol. Biotechnol Adv. https://doi.org/10.1016/j.biotechadv.2011.05.0
Li MW, Lang XY, Marcos MC et al (2021) CRISPR-mediated multigene integration enables Shikimate pathway refactoring for enhanced 2-phenylethanol biosynthesis in Kluyveromyces marxianus. Biotechnol Biofuels. https://doi.org/10.1186/s13068-020-01852-3
Zhu LH, Wang JH, Xu S et al (2021) Improved aromatic alcohol production by strengthening the shikimate pathway in Saccharomyces cerevisiae. Process Biochem. https://doi.org/10.1016/j.procbio.2021.01.025
Gu Y, Ma JB, Zhu YL et al (2020) Refactoring Ehrlich pathway for high-yield 2-phenylethanol production in Yarrowia lipolytica. ACS Synth Biol. https://doi.org/10.1021/acssynbio.9b00468
Braus GH (1991) Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol Rev. https://doi.org/10.1128/MMBR.55.3.349-370.1991
Kim B, Cho BR, Hahn JS (2014) Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via Ehrlich pathway. Biotechnol Bioeng. https://doi.org/10.1128/MMBR.55.3.349-370.1991
Daniel AS, Mónica SC, Carlos AS (2014) L-Phenylalanine Transport in Saccharomyces cerevisiae: Participation of GAP1, BAP2, and AGP1. J Amino Acids. https://doi.org/10.1155/2014/283962
Jolanta M, Katarzyna D, Karolina C et al (2019) Hydrolyzed corn stover as a promising feedstock for 2-phenylethanol production by nonconventional yeast. J Chem Technol Biotechnol. https://doi.org/10.1002/jctb.5823
Xu ZW, Chen Z, Lin LC, et al. (2021) Construction of CRISPR/CAS9 system for industrial Saccharomyces cerevisiae strain and genetic manipulation effect on 2-phenylethanol pathway. https://doi.org/10.21203/rs.3.rs-364472/v1
Zhang HB, Cao ML, Jiang XL, et al. (2014) De-novo synthesis of 2-phenylethanol by Enterobactersp. CGMCC 5087. BMC biotechnology. https://doi.org/10.1186/1472-6750-14-30
Mózsik L, Hoekzema M, de Kok NA et al (2021) CRISPR-based transcriptional activation tool for silent genes in filamentous fungi. Sci Rep. https://doi.org/10.1101/2020.10.13.338012
Gilbert LA, Larson MH, Morsut L et al (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. https://doi.org/10.1016/j.cell.2013.06.044
Alejandro C, Jonathan S, Suhani V et al (2015) Highly efficient Cas9-mediated transcriptional programming. Nat Methods. https://doi.org/10.1101/012880
Ni JP, Zhang GL, Qin L et al (2019) Simultaneously down-regulation of multiplex branch pathways using CRISPRi and fermentation optimization for enhancingβ-amyrin production in Saccharomyces cerevisiae. Synthetic and Systems Biotechnology. https://doi.org/10.1016/j.synbio.2019.02.002
Lian J, HamediRad M, Hu S et al (2017) Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat Commun. https://doi.org/10.1038/s41467-017-01695-x
Maximilian H, Kai S, Hélène E et al (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. https://doi.org/10.1186/s13059-016-1012-2
Kornel L, Tessa GM, Maximilian K et al (2019) CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz365
Laughery MF, Hunter T, Brown A et al (2015) New vectors for simple and streamlined CRISPR–Cas9 genome editing in Saccharomyces cerevisiae. Yeast. https://doi.org/10.1002/yea.3098
Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. https://doi.org/10.1016/S0076-6879(02)50957-5
Pigeau GM, Inglis DL (2010) Response of wine yeast (Saccharomyces cerevisiae) aldehyde dehydrogenases to acetaldehyde stress during Icewine fermentation. J Appl Microbiol. https://doi.org/10.1111/j.1365-2672.2007.03381.x
Qi LS, Larson MH, Gilbert LA et al (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. https://doi.org/10.1016/j.cell.2013.02.022
Marie FLR, Qi LS (2015) The New State of the Art: Cas9 for Gene Activation and Repression. Mol Cell Biol. https://doi.org/10.1128/MCB.00512-15
Xu H, **ao TF, Chen CH et al (2015) Sequence determinants of improved CRISPR sgRNA design. Genome Res. https://doi.org/10.1101/gr.191452.115
Yang YK, Liu GQ, Chen X et al (2020) High efficiency CRISPR/Cas9 genome editing system with an eliminable episomal sgRNA plasmid in Pichia pastoris. Enzyme Microb Technol. https://doi.org/10.1016/j.enzmictec.2020.109556
Kleinstiver BP, Pattanayak V, Prew MS et al (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. https://doi.org/10.1038/nature16526
Chen JS, Dagdas YS, Kleinstiver BP et al (2018) Enhanced Proofreading Governs CRISPR-Cas9 Targeting Accuracy. Biophys J. https://doi.org/10.1016/j.bpj.2017.11.1082
Yan M, Zhou SR, Xue HW (2015) CRISPR Primer Designer:Design primers for knockout and chromosome imaging CRISPR-Cas system. Journal of Integrative Plant Biology. CNKI:SUN:ZWXB.0.2015-07-002
Lian JZ, Carl S, Cao MF et al (2019) Multi-functional genome-wide CRISPR system for high throughput genotype–phenotype map**. Nat Commun. https://doi.org/10.1038/s41467-019-13621-4
Cordente AG, Solomon M, Alex S et al (2018) Novel wine yeast with ARO4 and TYR1 mutations that overproduce ‘floral’ aroma compounds 2-phenylethanol and 2-phenylethyl acetate. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-018-9054-x
Hassing EJ, de Groot PA, Marquenie VR et al (2019) Connecting central carbon and aromatic amino acid metabolisms to improve de novo 2-phenylethanol production in Saccharomyces cerevisiae. Metab Eng. https://doi.org/10.1016/j.ymben.2019.09.011
Pirkov I, Norbeck J, Gustafsson L et al (2008) A complete inventory of all enzymes in the eukaryotic methionine salvage pathway. FEBS J. https://doi.org/10.1111/j.1742-4658.2008.06552.x
Wang ZY, Bai XJ, Guo XN et al (2017) Regulation of crucial enzymes and transcription factors on 2-phenylethanol biosynthesis via Ehrlich pathway in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol. https://doi.org/10.1007/s10295-016-1852-5
Shen L, Nishimura YY, Matsuda F et al (2016) Overexpressing enzymes of the Ehrlich pathway and deleting genes of the competing pathway in Saccharomyces cerevisiae for increasing 2-phenylethanol production from glucose. J Biosci Bioeng. https://doi.org/10.1016/j.jbiosc.2015.12.022
Romagnoli G, Knijnenburg TA, Liti G et al (2015) Deletion of the Saccharomyces cerevisiae ARO8 gene, encoding an aromatic amino acid transaminase, enhances phenylethanol production from glucose. Yeast. https://doi.org/10.1002/yea.3015
Deed RC, Hou RY, Kinzurik MI et al (2019) The role of yeast ARO8, ARO9 and ARO10 genes in the biosynthesis of 3-(methylthio)-1-propanol from L-methionine during fermentation in synthetic grape medium. FEMS Yeast Res. https://doi.org/10.1093/femsyr/foy109
Xu ZW, Lin LC, Chen Z et al (2022) The same genetic regulation strategy produces inconsistent effects in different Saccharomyces cerevisiae strains for 2-phenylethanol production. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-022-11993-0
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fang, S., Fan, X., Li, J. et al. Triple-CRISPRi-mediated down-regulation of the shikimate pathway branch genes for enhancing 2-PE biosynthesis in Saccharomyces cerevisiae. Eur Food Res Technol (2024). https://doi.org/10.1007/s00217-023-04461-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00217-023-04461-0