Abstract
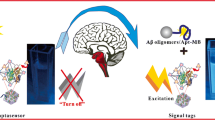
Aβ oligomers (AβO) are a dominant biomarker for early Alzheimer’s disease diagnosis. A fluorescent aptasensor coupled with conformational switch–induced hybridization was established to detect AβO. The fluorescent aptasensor is based on the interaction of fluorophore-labeled AβO-specific aptamer (FAM-Apt) against its partly complementary DNA sequence on the surface of magnetic beads (cDNA-MBs). Once the FAM-Apt binds to AβO, the conformational switch of FAM-Apt increases the tendency to be captured by cDNA-MBs. This causes a descending fluorescence of supernatant, which can be utilized to determine the levels of AβO. Thus, the base-pair matching above 12 between FAM-Apt and cDNA-MBs with increasing hybridizing free energies reached the ascending fluorescent signal equilibrium. The optimized aptasensor showed linearity from 1.7 ng mL−1 to 85.1 (R = 0.9977) with good recoveries (79.27–109.17%) in plasma. Furthermore, the established aptasensor possesses rational selectivity in the presence of monomeric Aβ, fibrotic Aβ, and interferences. Therefore, the developed aptasensor is capable of quantifying AβO in human plasma and possesses the potential to apply in clinical cases.
Graphical Abstract







Similar content being viewed by others
Abbreviations
- AβO:
-
Amyloid beta peptides
- AD:
-
Alzheimer’s disease
- FAM-Apt:
-
Fluorophore-labeled AβO-specific aptamer
- cDNA-MBs:
-
Complementary DNA sequence on the surface of magnetic beads
- CSF:
-
Cerebrospinal fluid
- HFIP:
-
1,1,1,3,3,3-Hexafluoro-2-propanol
- BSA:
-
Bovine serum albumin
- HSA:
-
Human serum albumin
- PTA:
-
Phosphotungstic acid
- SDS:
-
Sodium dodecyl sulfate
- Tris:
-
Tris(hydroxymethyl)aminomethane
- CE:
-
Capillary electrophoresis
- PDA:
-
Photodiode array
- BGE:
-
Background electrolyte
References
Ali GC, Guerchet M, Wu YT, Prince M, Prina M. The global prevalence of dementia. In: Prince M, Wimo A, Guerchet M, Ali GC, Wu YT, Prina M, editors. World Alzheimer Report 2015: the global impact of dementia: an analysis of prevalence, incidence, cost and trends. London: Alzheimer’s Disease International; 2015. pp. 10–28.
Guerchet M, Prince M, Prina M. Numbers of people with dementia around the world. Alzheimer’s Disease International; 2020. https://www.alzint.org/resource/numbers-of-people-with-dementia-worldwide/.
Burns A, Iliffe S. Alzheimer’s disease. BMJ. 2009;338: b158. https://doi.org/10.1136/bmj.b158.
Alzheimer’s disease facts and figures. Alzheimers Dement. 2021;17(3):327–406. https://doi.org/10.1002/alz.12328.
Cline EN, Bicca MA, Viola KL, Klein WL. The amyloid-beta oligomer hypothesis: beginning of the third decade. J Alzheimer’s Dis. 2018;64:S567-610. https://doi.org/10.3233/JAD-179941.
Braidy N, Zarka M, Jugder BE, Welch J, Jayasena T, Chan DKY, et al. The precursor to glutathione (GSH), γ-glutamylcysteine (GGC), can ameliorate oxidative damage and neuroinflammation induced by Aβ40 oligomers in human astrocytes. Front Aging Neurosci. 2019;11:177. https://doi.org/10.3389/fnagi.2019.00177.
Gao CM, Yam AY, Wang X, Magdangal E, Salisbury C, Peretz D, et al. Aβ40 oligomers identified as a potential biomarker for the diagnosis of Alzheimer’s disease. PLoS ONE. 2010;5(12): e15725. https://doi.org/10.1371/journal.pone.0015725.
Zhou L, Chan KH, Chu LW, Kwan JSC, Song YQ, Chen LH, et al. Plasma amyloid-β oligomers level is a biomarker for Alzheimer’s disease diagnosis. Biochem Biophys Res Commun. 2012;423(4):697–702. https://doi.org/10.1016/j.bbrc.2012.06.017.
Deane R, Wu Z, Zlokovic BV. RAGE (Yin) versus LRP (Yang) balance regulates Alzheimer amyloid β-peptide clearance through transport across the blood-brain barrier. Stroke. 2004;35(11 SUPPL. 1):2628–31. https://doi.org/10.1161/01.STR.0000143452.85382.d1.
Donahue JE, Flaherty SL, Johanson CE, Duncan JA, Silverberg GD, Miller MC, et al. RAGE, LRP-1, and amyloid-beta protein in Alzheimer’s disease. Acta Neuropathol. 2006;112(4):405–15. https://doi.org/10.1007/s00401-006-0115-3.
Nakamura A, Kaneko N, Villemagne VL, Kato T, Doecke J, Doré V, et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature. 2018;554(7691):249–54. https://doi.org/10.1038/nature25456.
Chiu MJ, Chen TF, Hu CJ, Yan SH, Sun Y, Liu BH, et al. Nanoparticle-based immunomagnetic assay of plasma biomarkers for differentiating dementia and prodromal states of Alzheimer’s disease: a cross-validation study. Nanomedicine. 2020;28: 102182. https://doi.org/10.1016/j.nano.2020.102182.
Youn YC, Lee BS, Kim GJ, Ryu JS, Lim K, Lee R, et al. Blood amyloid-β oligomerization as a biomarker of Alzheimer’s disease: a blinded validation study. J Alzheimer’s Dis. 2020;75(2):493–9. https://doi.org/10.3233/jad-200061.
Wang MJ, Yi S, Han JY, Park SY, Jang JW, Chun IK, et al. Oligomeric forms of amyloid-β protein in plasma as a potential blood-based biomarker for Alzheimer’s disease. Alzheimer’s Res Ther. 2017;9(1):98. https://doi.org/10.1186/s13195-017-0324-0.
Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–10. https://doi.org/10.1126/science.2200121.
Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–22. https://doi.org/10.1038/346818a0.
K. Tannenberg R, Al. Shamaileh H, H. Lauridsen L, R. Kanwar J, R. Dodd P, N. Veedu R. Nucleic acid aptamers as novel class of therapeutics to mitigate Alzheimer’s disease pathology. Curr Alzheimer Res. 2013;10(4):442–8. https://doi.org/10.2174/1567205011310040009.
Yan SR, Foroughi MM, Safaei M, Jahani S, Ebrahimpour N, Borhani F, et al. A review: Recent advances in ultrasensitive and highly specific recognition aptasensors with various detection strategies. Int J Biol Macromol. 2020;155:184–207. https://doi.org/10.1016/j.ijbiomac.2020.03.173.
Chen CH, Wang CC, Ko PY, Chen YL. Nanomaterial-based adsorbents and optical sensors for illicit drug analysis. J Food Drug Anal. 2020;28(4):655–76. https://doi.org/10.38212/2224-6614.1137.
Deng C, Liu H, Zhang M, Deng H, Lei C, Shen L, et al. Light-up nonthiolated aptasensor for low-mass, soluble amyloid-β40 oligomers at high salt concentrations. Anal Chem. 2018;90(3):1710–7. https://doi.org/10.1021/acs.analchem.7b03468.
Zhu X, Zhang N, Zhang Y, Liu B, Chang Z, Zhou Y, et al. A sensitive gold nanoparticle-based aptasensor for colorimetric detection of Aβ1-40 oligomers. Anal Methods. 2018;10(6):641–5. https://doi.org/10.1039/c7ay02918g.
You M, Yang S, An Y, Zhang F, He P. A novel electrochemical biosensor with molecularly imprinted polymers and aptamer-based sandwich assay for determining amyloid-β oligomer. J Electroanal Chem. 2020;862: 114017. https://doi.org/10.1016/j.jelechem.2020.114017.
Zhang Y, Figueroa-Miranda G, Lyu Z, Zafiu C, Willbold D, Offenhäusser A, et al. Monitoring amyloid-β proteins aggregation based on label-free aptasensor. Sens Actuators B Chem. 2019;288:535–42. https://doi.org/10.1016/j.snb.2019.03.049.
Chen W, Gao G, ** Y, Deng C. A facile biosensor for Aβ40O based on fluorescence quenching of prussian blue nanoparticles. Talanta. 2020;216: 120930. https://doi.org/10.1016/j.talanta.2020.120930.
Zhao Y, Li X, Yang Y, Si S, Deng C, Wu H. A simple aptasensor for Aβ40 oligomers based on tunable mismatched base pairs of dsDNA and graphene oxide. Biosens Bioelectron. 2020;149: 111840. https://doi.org/10.1016/j.bios.2019.111840.
Liu L, Chang Y, Yu J, Jiang M, **a N. Two-in-one polydopamine nanospheres for fluorescent determination of beta-amyloid oligomers and inhibition of beta-amyloid aggregation. Sens Actuators B Chem. 2017;251:359–65. https://doi.org/10.1016/j.snb.2017.05.106.
Zhu L, Zhang J, Wang F, Wang Y, Lu L, Feng C, et al. Selective amyloid β oligomer assay based on abasic site-containing molecular beacon and enzyme-free amplification. Biosens Bioelectron. 2016;78:206–12. https://doi.org/10.1016/j.bios.2015.11.048.
Tsukakoshi K, Abe K, Sode K, Ikebukuro K. Selection of DNA aptamers that recognize α-synuclein oligomers using a competitive screening method. Anal Chem. 2012;84(13):5542–7. https://doi.org/10.1021/ac300330g.
Liu F, Zhang JZH, Mei Y. The origin of the cooperativity in the streptavidin-biotin system: a computational investigation through molecular dynamics simulations. Sci Rep. 2016;6:1–11. https://doi.org/10.1038/srep27190.
Fezoui Y, Hartley DM, Harper JD, Khurana R, Walsh DM, Condron MM, et al. An improved method of preparing the amyloid β-protein for fibrillogenesis and neurotoxicity experiments. Amyloid. 2000;7(3):166–78. https://doi.org/10.3109/13506120009146831.
Bartolini M, Bertucci C, Bolognesi ML, Cavalli A, Melchiorre C, Andrisano V. Insight into the kinetic of amyloid β (1–42) peptide self-aggregation: elucidation of inhibitors’ mechanism of action. ChemBioChem. 2007;8(17):2152–61. https://doi.org/10.1002/cbic.200700427.
Bisceglia F, Natalello A, Serafini MM, Colombo R, Verga L, Lanni C, et al. An integrated strategy to correlate aggregation state, structure and toxicity of Aß 1–42 oligomers. Talanta. 2018;188:17–26. https://doi.org/10.1016/j.talanta.2018.05.062.
Gras SL, Waddington LJ, Goldie KN. Transmission electron microscopy of amyloid fibrils. In: Hill AF, Barnham KJ, Bottomley SP, Cappai R, editors. Protein folding, misfolding, and disease. 2011. p. 197–214. https://doi.org/10.1007/978-1-60327-223-0_13
Ryan TM, Caine J, Mertens HDT, Kirby N, Nigro J, Breheney K, et al. Ammonium hydroxide treatment of Aβ produces an aggregate free solution suitable for biophysical and cell culture characterization. PeerJ. 2013;1: e73. https://doi.org/10.7717/peerj.73.
Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–9. https://doi.org/10.1126/science.1079469.
Hoshi M, Sato M, Matsumoto S, Noguchi A, Yasutake K, Yoshida N, et al. Spherical aggregates of β-amyloid (amylospheroid) show high neurotoxicity and activate tau protein kinase I/glycogen synthase kinase-3β. Proc Natl Acad Sci USA. 2003;100(11):6370–5. https://doi.org/10.1073/pnas.1237107100.
Jan A, Hartley DM, Lashuel HA. Preparation and characterization of toxic Aβ aggregates for structural and functional studies in Alzheimer’s disease research. Nat Protoc. 2010;5(6):1186–209. https://doi.org/10.1038/nprot.2010.72.
Parkinson GN. Fundamentals of quadruplex structures. In: Neidle S, Balasubramanian S, editors. Quadruplex nucleic acids. 2007. p. 1–30. https://doi.org/10.1039/9781847555298-00001
Hou MH, Lin SB, Yuann JMP, Lin WC, Wang AHJ, Kan LS. Effects of polyamines on the thermal stability and formation kinetics of DNA duplexes with abnormal structure. Nucleic Acids Res. 2001;29(24):5121–8. https://doi.org/10.1093/nar/29.24.5121.
Zhang Z, Huang W, Tang J, Wang E, Dong S. Conformational transition of DNA induced by cationic lipid vesicle in acidic solution: spectroscopy investigation. Biophys Chem. 2002;97(1):7–16. https://doi.org/10.1016/S0301-4622(02)00006-6.
Lomzov AA, Vorobjev YN, Pyshnyi DV. Evaluation of the Gibbs free energy changes and melting temperatures of DNA/DNA duplexes using hybridization enthalpy calculated by molecular dynamics simulation. J Phys Chem B. 2015;119(49):15221–34. https://doi.org/10.1021/acs.jpcb.5b09645.
Largy E, Mergny JL, Gabelica V. Role of alkali metal ions in G-quadruplex nucleic acid structure and stability. In: Sigel A, Sigel H, Sigel RKO, editors. Met Ions Life Sci. 2016. p. 203–58. https://doi.org/10.1007/978-3-319-21756-7_7
Cai S, Yan J, **ong H, Liu Y, Peng D, Liu Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst. 2018;143(22):5317–38. https://doi.org/10.1039/c8an01467a.
Bhattacharyya D, Arachchilage GM, Basu S. Metal cations in G-quadruplex folding and stability. Front Chem. 2016;4:38. https://doi.org/10.3389/fchem.2016.00038.
Simonsson T. G-Quadruplex DNA structures variations on a theme. Biol Chem. 2001;382(4):621–8. https://doi.org/10.1515/BC.2001.073.
Shrivastava A, Gupta V. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron Young Sci. 2011;2(1):21–5. https://doi.org/10.4103/2229-5186.79345.
Acknowledgements
We gratefully acknowledge the support of the Ministry of Science and Technology of Taiwan (MOST 110-2113-M-194-007).
The application of this developed aptasensor to human plasma acquired from healthy subjects in Kaohsiung Medical University was approved by the Institutional Review Board (IRB) of Kaohsiung Medical University Chung-Ho Memorial Hospital (KMUHIRB-E(I)-20190389).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, CH., Jong, YJ., Chao, YY. et al. Fluorescent aptasensor based on conformational switch–induced hybridization for facile detection of β-amyloid oligomers. Anal Bioanal Chem 414, 8155–8165 (2022). https://doi.org/10.1007/s00216-022-04350-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-04350-7