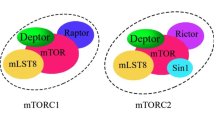
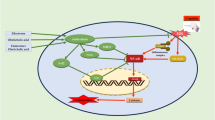
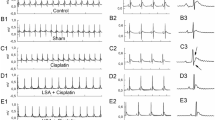
Abstract
Cisplatin is a widely used chemotherapeutic agent for the treatment of various cancers. However, the clinical use of cisplatin is limited by its cardiotoxic side effects. The primary mechanisms implicated in this cardiotoxicity include mitochondrial dysfunction, oxidative stress, inflammation, and apoptotic. Numerous natural compounds (NCs) have been introduced as promising protective factors against cisplatin-mediated cardiac damage. The current review summarized the potential of various NCs as cardioprotective agents at the molecular levels. These compounds exhibited potent antioxidant and anti-inflammatory effects by interaction with the PI3K/AKT, AMPK, Nrf2, NF-κB, and NLRP3/caspase-1/GSDMD pathways. Generally, the modulation of these signaling pathways by NCs represents a promising strategy for improving the therapeutic index of cisplatin by reducing its cardiac side effects.


Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
Abbreviations
- ACC:
-
Acetyl-CoA carboxylase
- ALA:
-
Alpha-lipoic acid
- AMPK:
-
AMP-activated protein kinase
- Apaf-1:
-
Apoptosis-protease activating factor 1
- APO:
-
Apocynin
- ARE:
-
Antioxidant response element
- BACH1:
-
BTB and CNC homology 1
- BAX:
-
Bcl-2-associated X protein
- Bcl-2:
-
B-cell lymphoma-2
- Bzip:
-
Basic leucine zipper
- CAT:
-
Catalase
- COX-2:
-
Cyclooxygenase-2
- CNC:
-
Cap ‘n’ collar
- CUR:
-
Curcumin
- CYN:
-
Cyaniding
- DUBs:
-
Deubiquitinases
- E1:
-
Ubiquitin-activating enzymes
- E2:
-
Ubiquitin-conjugating enzymes
- E3:
-
Ubiquitin ligases
- ELA:
-
Ellagic acid
- ER:
-
Endoplasmic reticulum
- ETC:
-
Electron transport chain
- GCL:
-
Glutamate cysteine ligase
- GPx:
-
Glutathione peroxidase
- GSDMD:
-
Gasdermin D
- GSH:
-
Glutathione
- GSK-3β:
-
Glycogen synthase kinase-3 beta
- GSTs:
-
Glutathione S-transferases
- HES:
-
Hesperidin
- HO-1:
-
Heme oxygenase-1
- ICA:
-
Icariin
- IKK:
-
IκB kinase
- IκB-α:
-
NF-κB inhibitor-alpha
- ILs:
-
Interleukins
- Inos:
-
Inducible nitric oxide synthase
- I/R:
-
Ischemia/reperfusion
- IRF3:
-
Interferon regulatory factor 3
- Keap1:
-
Kelch-like ECH-associated protein 1
- KPF:
-
Kaempferol
- LUT:
-
Luteolin
- LYC:
-
Lycopene
- Maf:
-
Small musculoaponeurotic fibrosarcoma oncogene
- MAL:
-
Maltol
- MCP-1:
-
Monocyte chemotactic protein-1
- MDA:
-
Malondialdehyde
- MOR:
-
Morphine
- NAR:
-
Naringin
- NCs:
-
Natural compounds
- NEK7:
-
NIMA-related kinase 7
- NF-Κb:
-
Nuclear factor-kappa B
- NLRP3:
-
NOD-like receptor family pyrin domain-containing 3
- NQO-1:
-
NAD(P)H quinone oxidoreductase-1
- Nrf2:
-
Nuclear factor erythroid 2-related factors
- p38 MAPK:
-
P38 mitogen-activated protein kinase
- PI3Ks:
-
Phosphoinositide 3-kinases
- PIP2:
-
Phosphatidylinositol (4,5)-bisphosphate
- PIP3:
-
Phosphatidylinositol (3,4,5)-trisphosphate
- QUE:
-
Quercetin
- ROS:
-
Reactive oxygen species
- RSV:
-
Resveratrol
- RUT:
-
Rutin trihydrate
- SA:
-
Sinapic acid
- SalB:
-
Salvianolic acid B
- SAPs:
-
Saponins
- SOD:
-
Superoxide dismutase
- STING:
-
Stimulator of interferon genes
- SYL:
-
Silymarin
- TBK1:
-
TANK-binding kinase 1
- TG:
-
Tangeretin
- TNF-α:
-
Tumor necrosis factor alpha
- TQ:
-
Thymoquinone
- TSC2:
-
Tuberous sclerosis complex 2
- WOG:
-
Wogonin
- ZN:
-
Zingerone
References
Abudalo R, Gammoh O, Altaber S et al (2024) Mitigation of cisplatin-induced cardiotoxicity by Isorhamnetin: mechanistic insights into oxidative stress, inflammation, and apoptosis modulation. Toxicol Reports 12:564–573. https://doi.org/10.1016/j.toxrep.2024.05.003
Adalı F, Gonul Y, Kocak A et al (2016) Effects of thymoquinone against cisplatin-induced cardiac injury in rats. Acta Cir Bras 31:271–277. https://doi.org/10.1590/S0102-865020160040000008
Ahn H, Lee G, Han B-C et al (2022) Maltol, a natural flavor enhancer, inhibits NLRP3 and non-canonical inflammasome activation. Antioxidants 11:1923. https://doi.org/10.3390/antiox11101923
Akhtar MN, Saeed R, Saeed F et al (2023) Silymarin: a review on paving the way towards promising pharmacological agent. Int J Food Prop 26:2256–2272
Al-Hussaniy HA, Alburghaif AH, Alkhafaje Z et al (2023) Chemotherapy-induced cardiotoxicity: a new perspective on the role of Digoxin, ATG7 activators, Resveratrol, and herbal drugs. J Med Life 16:491–500. https://doi.org/10.25122/jml-2022-0322
Amador-Martínez I, Hernández-Cruz EY, Jiménez-Uribe AP et al (2021) Mitochondrial transplantation: is it a feasible therapy to prevent the cardiorenal side effects of cisplatin? Futur Pharmacol 1:3–26
Antonia RJ, Hagan RS, Baldwin AS (2021) Expanding the view of IKK: new substrates and new biology. Trends Cell Biol 31:166–178. https://doi.org/10.1016/j.tcb.2020.12.003
Arafa E-SA, Shurrab NT, Buabeid MA (2021) Therapeutic implications of a polymethoxylated flavone, tangeretin, in the management of cancer via modulation of different molecular pathways. Adv Pharmacol Pharm Sci 2021:4709818. https://doi.org/10.1155/2021/4709818
Arbeláez LFG, Pardo AC, Fantinelli JC et al (2018) Cardioprotection and natural polyphenols: an update of clinical and experimental studies. Food Funct 9:6129–6145
Ardianto C, Budiatin AS, Sumartha INB et al (2021) Resveratrol ameliorates physical and psychological stress-induced depressive-like behavior. J Basic Clin Physiol Pharmacol 32:335–340
Baird L, Yamamoto M (2020) The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol Cell Biol 40:e00099-e120. https://doi.org/10.1128/MCB.00099-20
Bangar SP, Chaudhary V, Sharma N et al (2023) Kaempferol: a flavonoid with wider biological activities and its applications. Crit Rev Food Sci Nutr 63:9580–9604
Bard JAM, Goodall EA, Greene ER et al (2018) Structure and function of the 26S proteasome. Annu Rev Biochem 87:697–724. https://doi.org/10.1146/annurev-biochem-062917-011931
Batiha GE-S, Beshbishy AM, Ikram M et al (2020) The pharmacological activity, biochemical properties, and pharmacokinetics of the major natural polyphenolic flavonoid: quercetin. Foods 9:374. https://doi.org/10.3390/foods9030374
Bayazid AB, Chun EM, Al Mijan M et al (2021) Anthocyanins profiling of bilberry (Vaccinium myrtillus L.) extract that elucidates antioxidant and anti-inflammatory effects. Food Agric Immunol 32:713–726
Beurel E, Grieco SF, Jope RS (2015) Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol Ther 148:114–131. https://doi.org/10.1016/j.pharmthera.2014.11.016
Boarescu P-M, Chirilă I, Bulboacă AE et al (2019) Effects of curcumin nanoparticles in isoproterenol-induced myocardial infarction. Oxid Med Cell Longev 2019:7847142. https://doi.org/10.1155/2019/7847142
Brown A, Kumar S, Tchounwou PB (2019) Cisplatin-based chemotherapy of human cancers. J Cancer Sci Ther 11:97
Bukhari IA, Mohamed OY, Alhowikan AM et al (2022) Protective effect of Rutin trihydrate against dose-dependent, cisplatin-induced cardiac toxicity in isolated perfused rat’s heart. Cureus 14:e21572. https://doi.org/10.7759/cureus.21572
Camini FC, Costa DC (2020) Silymarin: Not just another antioxidant. J Basic Clin Physiol Pharmacol 31:20190206
Cantrell DA (2001) Phosphoinositide 3-kinase signalling pathways. J Cell Sci 114:1439–1445. https://doi.org/10.1242/jcs.114.8.1439
Caseiro M, Ascenso A, Costa A et al (2020) Lycopene in human health. LWT 127:109323. https://doi.org/10.1016/j.lwt.2020.109323
Choudhary N, Bawari S, Burcher JT et al (2023) Targeting cell signaling pathways in lung cancer by bioactive phytocompounds. Cancers (Basel) 15:3980
Cockram PE, Kist M, Prakash S et al (2021) Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ 28:591–605. https://doi.org/10.1038/s41418-020-00708-5
Costa BM, Mengal V, Brasil GA et al (2022) Ellagic acid prevents myocardial infarction-induced left ventricular diastolic dysfunction in ovariectomized rats. J Nutr Biochem 105:108990
Darakhshan S, Pour AB, Colagar AH, Sisakhtnezhad S (2015) Thymoquinone and its therapeutic potentials. Pharmacol Res 95:138–158
Dastani M, Bigdelu L, Hoseinzadeh M et al (2019) The effects of curcumin on the prevention of atrial and ventricular arrhythmias and heart failure in patients with unstable angina: a randomized clinical trial. Avicenna J Phytomed 9:1
Dugbartey GJ, Peppone LJ, de Graaf IAM (2016) An integrative view of cisplatin-induced renal and cardiac toxicities: molecular mechanisms, current treatment challenges and potential protective measures. Toxicology 371:58–66. https://doi.org/10.1016/j.tox.2016.10.001
El-Awady E-SE, Moustafa YM, Abo-Elmatty DM, Radwan A (2011) Cisplatin-induced cardiotoxicity Mechanisms and cardioprotective strategies. Eur J Pharmacol 650:335–341. https://doi.org/10.1016/j.ejphar.2010.09.085
El-Sawalhi MM, Ahmed LA (2014) Exploring the protective role of apocynin, a specific NADPH oxidase inhibitor, in cisplatin-induced cardiotoxicity in rats. Chem Biol Interact 207:58–66. https://doi.org/10.1016/j.cbi.2013.11.008
Elsayed A, Elkomy A, Alkafafy M et al (2022) Ameliorating effect of lycopene and N-Acetylcysteine against cisplatin-induced cardiac injury in rats. Pak Vet J 42:107–111. https://doi.org/10.29261/pakvetj/2021.035
El-Sheikh AAK, Khired Z (2021) Morphine deteriorates cisplatin-induced cardiotoxicity in rats and induces dose-dependent cisplatin chemoresistance in MCF-7 human breast cancer cells. Cardiovasc Toxicol 21:553–562. https://doi.org/10.1007/s12012-021-09646-1
Farkhondeh T, Samarghandian S, Borji A (2017) An overview on cardioprotective and anti-diabetic effects of thymoquinone. Asian Pac J Trop Med 10:849–854. https://doi.org/10.1016/j.apjtm.2017.08.020
Gelen V, Şengül E (2020) Antioxidant, anti-inflammatory and antiapoptotic effects of Naringin on cardiac damage induced by cisplatin. Indian J Tradit Knowl 19:459–465. https://doi.org/10.56042/ijtk.v19i2.35371
Gimenes NC, Silveira E, Tambourgi EB (2021) An overview of proteases: production, downstream processes and industrial applications. Sep Purif Rev 50:223–243. https://doi.org/10.1080/15422119.2019.1677249
González A, García-Gómez-Heras S, Franco-Rodríguez R et al (2023) Cisplatin cycles treatment sustains cardiovascular and renal damage involving TLR4 and NLRP3 pathways. Pharmacol Res Perspect 11:e01108
Hassanizadeh S, Shojaei M, Bagherniya M et al (2023) Effect of nano-curcumin on various diseases: a comprehensive review of clinical trials. Biofactors 49:512–533. https://doi.org/10.1002/biof.1932
He G, Chen G, Liu W et al (2023) Salvianolic acid B: a review of pharmacological effects, safety, combination therapy, new dosage forms, and novel drug delivery routes. Pharmaceutics 15:2235
Hemalatha KL, Prince PSM (2016) Preventive effects of zingerone on cardiac mitochondrial oxidative stress, calcium ion overload and adenosine triphosphate depletion in isoproterenol induced myocardial infarcted rats. RSC Adv 6:112332–112339
Hu Q-F, Sun A-J (2020) Cardioprotective effect of alpha-lipoic acid and its mechanisms. Cardiol plus 5:109–117
Hu H, Li Y, Yang Y et al (2022) Effect of a plateau environment on the oxidation state of the heart and liver through AMPK/p38 MAPK/Nrf2-ARE signaling pathways in Tibetan and DLY pigs. Animals 12:1219
Hussein A, Ahmed AAE, Shouman SA, Sharawy S (2012) Ameliorating effect of DL-α-lipoic acid against cisplatin-induced nephrotoxicity and cardiotoxicity in experimental animals. Drug Discov Ther 6:147–156
Inoki K, Li Y, Zhu T et al (2002) TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4:648–657. https://doi.org/10.1038/ncb839
Irfan Afridi M, Zheng Z, Liu J et al (2023) The bZIP transcription factor BATF3/ZIP-10 suppresses innate immunity by attenuating PMK-1/p38 signaling. Int Immunol 35:181–196. https://doi.org/10.1093/intimm/dxac053
Jara-Palacios MJ, Santisteban A, Gordillo B et al (2019) Comparative study of red berry pomaces (blueberry, red raspberry, red currant and blackberry) as source of antioxidants and pigments. Eur Food Res Technol 245:1–9. https://doi.org/10.1007/s00217-018-3135-z
Jia Y, Guo H, Cheng X et al (2022) Hesperidin protects against cisplatin-induced cardiotoxicity in mice by regulating the p62-Keap1-Nrf2 pathway. Food Funct 13:4205–4215. https://doi.org/10.1039/d2fo00298a
**drich K, Degnan BM (2016) The diversification of the basic leucine zipper family in eukaryotes correlates with the evolution of multicellularity. BMC Evol Biol 16:28. https://doi.org/10.1186/s12862-016-0598-z
Juan CA, Pérez de la Lastra JM, Plou FJ, Pérez-Lebeña E (2021) The chemistry of reactive oxygen species (ROS) revisited: outlining their role in biological macromolecules (DNA, lipids and proteins) and induced pathologies. Int J Mol Sci 22:4642
Juang Y-P, Liang P-H (2020) Biological and pharmacological effects of synthetic saponins. Molecules 25:4974
Kamisah Y, Jalil J, Yunos NM, Zainalabidin S (2023) Cardioprotective properties of kaempferol: a review. Plants 12:2096
Kannan MM, Quine SD (2013) Ellagic acid inhibits cardiac arrhythmias, hypertrophy and hyperlipidaemia during myocardial infarction in rats. Metabolism 62:52–61
Khadrawy YA, Hosny EN, El-Gizawy MM et al (2021) The effect of curcumin nanoparticles on cisplatin-induced cardiotoxicity in male Wistar albino rats. Cardiovasc Toxicol 21:433–443. https://doi.org/10.1007/s12012-021-09636-3
Khan S, Zhang D, Zhang Y et al (2016) Wogonin attenuates diabetic cardiomyopathy through its anti-inflammatory and anti-oxidative properties. Mol Cell Endocrinol 428:101–108
Khuntia G, Dash JR, Jena B et al (2023) Hesperidin attenuates arsenic trioxide-induced cardiac toxicity in rats. Asian Pac J Trop Biomed 13:156–164
Kuznetsov AV, Javadov S, Grimm M et al (2020) Crosstalk between mitochondria and cytoskeleton in cardiac cells. Cells 9:222
Kuzu M, Kandemir FM, Yıldırım S et al (2021) Attenuation of sodium arsenite-induced cardiotoxicity and neurotoxicity with the antioxidant, anti-inflammatory, and antiapoptotic effects of hesperidin. Environ Sci Pollut Res Int 28:10818–10831. https://doi.org/10.1007/s11356-020-11327-5
Lee Y-M, Cheng P-Y, Chen S-Y et al (2011) Wogonin suppresses arrhythmias, inflammatory responses, and apoptosis induced by myocardial ischemia/reperfusion in rats. J Cardiovasc Pharmacol 58:133–142
Lin Y, Jiang M, Chen W et al (2019) Cancer and ER stress: Mutual crosstalk between autophagy, oxidative stress and inflammatory response. Biomed Pharmacother 118:109249. https://doi.org/10.1016/j.biopha.2019.109249
Lin Z, Bao Y, Hong B et al (2021) Salvianolic acid B attenuated cisplatin-induced cardiac injury and oxidative stress via modulating Nrf2 signal pathway. J Toxicol Sci 46:199–207. https://doi.org/10.2131/jts.46.199
Liu Y, Wang J, Zhang C et al (2020) Tangeretin prevents cardiac failure induced by reperfusion/ischaemia by inhibiting apoptosis, endoplasmic reticulum stress, and JNK/ERK pathway. Arch Med Sci 16. https://doi.org/10.5114/aoms.2020.97052
Luan F, Rao Z, Peng L et al (2022) Cinnamic acid preserves against myocardial ischemia/reperfusion injury via suppression of NLRP3/Caspase-1/GSDMD signaling pathway. Phytomedicine 100:154047
Luo Y, Shang P, Li D (2017) Luteolin: a flavonoid that has multiple cardio-protective effects and its molecular mechanisms. Front Pharmacol 8:692
Luo F, Luo S, Wu Y (2021) Zingerone alleviates myocardial ischemia/reperfusion injury. Curr Top Nutraceutical Res 19:543. https://doi.org/10.37290/ctnr2641-452X.19:543-549
Ma X, Lu J, Gu X-R et al (2022) Cardioprotective effects and mechanisms of saponins on cardiovascular disease. Nat Prod Commun 17:1934578X221147404
Mi X, Hou J, Wang Z et al (2018) The protective effects of maltol on cisplatin-induced nephrotoxicity through the AMPK-mediated PI3K/Akt and p53 signaling pathways. Sci Rep 8:15922. https://doi.org/10.1038/s41598-018-34156-6
Miao J, Lin F, Huang N, Teng Y (2021) Improving anti-inflammatory effect of luteolin with nano-micelles in the bacteria-induced lung infection. J Biomed Nanotechnol 17:1229–1241
Mohammad A, Babiker F, Al-Bader M (2023) Effects of apocynin, a NADPH oxidase inhibitor, in the protection of the heart from ischemia/reperfusion injury. Pharmaceuticals 16:492. https://doi.org/10.3390/ph16040492
Nageeb MM, Saadawy SF, Attia SH (2022) Breast milk mesenchymal stem cells abate cisplatin-induced cardiotoxicity in adult male albino rats via modulating the AMPK pathway. Sci Rep 12. https://doi.org/10.1038/s41598-022-22095-2
Najafi N, Mehri S, Ghasemzadeh Rahbardar M, Hosseinzadeh H (2022) Effects of alpha lipoic acid on metabolic syndrome: a comprehensive review. Phyther Res 36:2300–2323. https://doi.org/10.1002/ptr.7406
Nam LB, Keum Y-S (2019) Binding partners of NRF2: Functions and regulatory mechanisms. Arch Biochem Biophys 678:108184. https://doi.org/10.1016/j.abb.2019.108184
Ngo V, Duennwald ML (2022) Nrf2 and oxidative stress: a general overview of mechanisms and implications in human disease. Antioxidants 11:2345. https://doi.org/10.3390/antiox11122345
Obeng E (2020) Apoptosis (programmed cell death) and its signals-a review. Brazilian J Biol 81:1133–1143
Oguzturk H, Ciftci O, Cetin A et al (2016) Beneficial effects of hesperidin following cis-diamminedichloroplatinum-induced damage in heart of rats. Niger J Clin Pract 19:99–103. https://doi.org/10.4103/1119-3077.173707
Pandey P, Khan F (2021) A mechanistic review of the anticancer potential of hesperidin, a natural flavonoid from citrus fruits. Nutr Res 92:21–31
Pandi A, Kalappan VM (2021) Pharmacological and therapeutic applications of Sinapic acid—an updated review. Mol Biol Rep 48:3733–3745
Pang Y, Xu X, **ang X et al (2021) High fat activates O-GlcNAcylation and affects AMPK/ACC pathway to regulate lipid metabolism. Nutrients 13:1740
Pyrzynska K (2022) Hesperidin: a review on extraction methods, stability and biological activities. Nutrients 14:2387
Qi Y, Ying Y, Zou J et al (2020) Kaempferol attenuated cisplatin-induced cardiac injury via inhibiting STING/NF-κB-mediated inflammation. Am J Transl Res 12:8007–8018
Qi Y, Fu S, Pei D et al (2022) Luteolin attenuated cisplatin-induced cardiac dysfunction and oxidative stress via modulation of Keap1/Nrf2 signaling pathway. Free Radic Res 56:209–221. https://doi.org/10.1080/10715762.2022.2067042
Qian P, Yan LJ, Li YQ et al (2018) Cyanidin ameliorates cisplatin-induced cardiotoxicity via inhibition of ROS-mediated apoptosis. Exp Ther Med 15:1959–1965. https://doi.org/10.3892/etm.2017.5617
Rahman S, Mathew S, Nair P et al (2021) Health benefits of cyanidin-3-glucoside as a potent modulator of Nrf2-mediated oxidative stress. Inflammopharmacology 29:907–923
Rajan P, Natraj P, Ranaweera SS et al (2022) Anti-diabetic effect of hesperidin on palmitate (PA)-treated HepG2 cells and high fat diet-induced obese mice. Food Res Int 162:112059
Rateb A, Gomaa A, Hussein OA (2022) Adipose tissue-mesenchymal stem cells improve cisplatin-induced cardiotoxicity in rats via modulating apoptosis, oxidative stress, inflammation, and angiogenesis. Egypt Acad J Biol Sci D Histol Histochem 14:119–145
Rathinaswamy MK, Burke JE (2020) Class I phosphoinositide 3-kinase (PI3K) regulatory subunits and their roles in signaling and disease. Adv Biol Regul 75:100657. https://doi.org/10.1016/j.jbior.2019.100657
Rezaee R, Sheidary A, Jangjoo S et al (2021) Cardioprotective effects of hesperidin on carbon monoxide poisoned in rats. Drug Chem Toxicol 44:668–673
Saleh RM, Awadin WF, Elseady YY, Waheish FE (2014) Renal and cardiovascular damage induced by cisplatin in rats. Life Sci J 11:191–203
Salehi B, Berkay Yılmaz Y, Antika G et al (2019) Insights on the use of α-lipoic acid for therapeutic purposes. Biomolecules 9. https://doi.org/10.3390/biom9080356
Sapian S, Taib IS, Katas H et al (2022) The role of anthocyanin in modulating diabetic cardiovascular disease and its potential to be developed as a nutraceutical. Pharmaceuticals (Basel) 15. https://doi.org/10.3390/ph15111344
Savla SR, Laddha AP, Kulkarni YA (2021) Pharmacology of apocynin: a natural acetophenone. Drug Metab Rev 53:542–562. https://doi.org/10.1080/03602532.2021.1895203
Semwal R, Joshi SK, Semwal RB, Semwal DK (2021) Health benefits and limitations of rutin - a natural flavonoid with high nutraceutical value. Phytochem Lett 46:119–128. https://doi.org/10.1016/j.phytol.2021.10.006
Sengoku T, Shiina M, Suzuki K et al (2022) Structural basis of transcription regulation by CNC family transcription factor, Nrf2. Nucleic Acids Res 50:12543–12557. https://doi.org/10.1093/nar/gkac1102
Serasanambati M, Chilakapati SR (2016) Function of nuclear factor kappa B (NF-kB) in human diseases-a review. South Indian J Biol Sci 2:368–387. https://doi.org/10.22205/sijbs/2016/v2/i4/103443
Sha J, Li J, Zhou Y et al (2021) The p53/p21/p16 and PI3K/Akt signaling pathways are involved in the ameliorative effects of maltol on D-galactose-induced liver and kidney aging and injury. Phyther Res 35:4411–4424. https://doi.org/10.1002/ptr.7142
Shahid F, Farooqui Z, Khan F (2018) Cisplatin-induced gastrointestinal toxicity: an update on possible mechanisms and on available gastroprotective strategies. Eur J Pharmacol 827:49–57
Shamsabadi S, Nazer Y, Ghasemi J et al (2023) Promising influences of zingerone against natural and chemical toxins: a comprehensive and mechanistic review. Toxicon 233:107247. https://doi.org/10.1016/j.toxicon.2023.107247
Sharifi-Rad J, Rodrigues CF, Sharopov F et al (2020) Diet, lifestyle and cardiovascular diseases: linking pathophysiology to cardioprotective effects of natural bioactive compounds. Int J Environ Res Public Health 17:2326
Sharifi-Rad J, Quispe C, Castillo CMS et al (2022) Ellagic acid: a review on its natural sources, chemical stability, and therapeutic potential. Oxid Med Cell Longev 2022:3848084. https://doi.org/10.1155/2022/3848084
Sharifi-Rad J, Herrera-Bravo J, Salazar LA et al (2021) The therapeutic potential of wogonin observed in preclinical studies. Evidence-Based Complement Altern Med 2021:9935451. https://doi.org/10.1155/2021/9935451
Shen R, Wang J-H (2018) The effect of icariin on immunity and its potential application. Am J Clin Exp Immunol 7:50–56
Shilpa VS, Shams R, Dash KK et al (2023) Phytochemical properties, extraction, and pharmacological benefits of naringin: a review. Molecules 28:5623
Singh B, Kumar A, Singh H et al (2020) Zingerone produces antidiabetic effects and attenuates diabetic nephropathy by reducing oxidative stress and overexpression of NF-κB, TNF-α, and COX-2 proteins in rats. J Funct Foods 74:104199
Soliman AF, Anees LM, Ibrahim DM (2018) Cardioprotective effect of zingerone against oxidative stress, inflammation, and apoptosis induced by cisplatin or gamma radiation in rats. Naunyn Schmiedebergs Arch Pharmacol 391:819–832. https://doi.org/10.1007/s00210-018-1506-4
Song Y-J, Zhong C-B, Wu W (2020) Resveratrol and diabetic cardiomyopathy: focusing on the protective signaling mechanisms. Oxid Med Cell Longev 2020:7051845. https://doi.org/10.1155/2020/7051845
Stojic IM, Zivkovic VI, Srejovic IM et al (2018) Cisplatin and cisplatin analogues perfusion through isolated rat heart: the effects of acute application on oxidative stress biomarkers. Mol Cell Biochem 439:19–33. https://doi.org/10.1007/s11010-017-3132-8
Tejaswini R, Kumar IP (2024) Neurotoxicity evaluation of rutin trihydrate vs. metformin in zebrafish larvae: a comparative risk assessment study. In: E3S Web of Conferences. https://doi.org/10.1051/e3sconf/202447700068
Tian C, Liu X, Chang Y et al (2021) Investigation of the anti-inflammatory and antioxidant activities of luteolin, kaempferol, apigenin and quercetin. South African J Bot 137:257–264
Vishwakarma A, Singh TU, Rungsung S et al (2018) Effect of kaempferol pretreatment on myocardial injury in rats. Cardiovasc Toxicol 18:312–328
Viswanatha GL, Shylaja H, Keni R et al (2022) A systematic review and meta-analysis on the cardio-protective activity of naringin based on pre-clinical evidences. Phyther Res 36:1064–1092. https://doi.org/10.1002/ptr.7368
Wang X, Robbins J (2014) Proteasomal and lysosomal protein degradation and heart disease. J Mol Cell Cardiol 71:16–24. https://doi.org/10.1016/j.yjmcc.2013.11.006
Wang J, He D, Zhang Q et al (2009) Resveratrol protects against Cisplatin-induced cardiotoxicity by alleviating oxidative damage. Cancer Biother Radiopharm 24:675–680. https://doi.org/10.1089/cbr.2009.0679
Wang S-H, Tsai K-L, Chou W-C et al (2022) Quercetin mitigates cisplatin-induced oxidative damage and apoptosis in cardiomyocytes through Nrf2/HO-1 signaling pathway. Am J Chin Med 50:1281–1298. https://doi.org/10.1142/S0192415X22500537
Wang J, Zhang R, Hu J et al (2024) Tangeretin, an active flavonoid in citrus peel, alleviates cisplatin-induced cardiotoxicity via the activation of AMPK and the prevention on mitochondrial dysfunction. PREPRINT (Version 1) available at Research Square. https://doi.org/10.21203/rs.3.rs-3868935/v1
**a S, Hollingsworth LR, Wu H (2020) Mechanism and regulation of gasdermin-mediated cell death. Cold Spring Harb Perspect Biol 12:a036400
**a J, Hu JN, Zhang RB et al (2022) Icariin exhibits protective effects on cisplatin-induced cardiotoxicity via ROS-mediated oxidative stress injury in vivo and in vitro. Phytomedicine 104. https://doi.org/10.1016/j.phymed.2022.154331
**ng J-J, Mi X-J, Hou J-G et al (2022) Maltol mitigates cisplatin-evoked cardiotoxicity via inhibiting the PI3K/Akt signaling pathway in rodents in vivo and in vitro. Phytother Res 36:1724–1735. https://doi.org/10.1002/ptr.7405
**ng J-J, Hou J-G, Liu Y, et al (2019) Supplementation of saponins from leaves of panax quinquefolius mitigates cisplatin-evoked cardiotoxicity via inhibiting oxidative stress-associated inflammation and apoptosis in mice. Antioxidants (Basel, Switzerland) 8. https://doi.org/10.3390/antiox8090347
Xu J, Zhang B, Chu Z et al (2021) Wogonin alleviates cisplatin-induced cardiotoxicity in mice via inhibiting gasdermin D-mediated pyroptosis. J Cardiovasc Pharmacol 78:597–603. https://doi.org/10.1097/FJC.0000000000001085
Yang D, Wang T, Long M, Li P (2020) Quercetin: its main pharmacological activity and potential application in clinical medicine. Oxid Med Cell Longev 2020:8825387. https://doi.org/10.1155/2020/8825387
Yao C, Veleva T, Scott L Jr et al (2018) Enhanced cardiomyocyte NLRP3 inflammasome signaling promotes atrial fibrillation. Circulation 138:2227–2242. https://doi.org/10.1161/CIRCULATIONAHA.118.035202
Yarmohammadi F, Wallace Hayes A, Najafi N, Karimi G (2020) The protective effect of natural compounds against rotenone-induced neurotoxicity. J Biochem Mol Toxicol. https://doi.org/10.1002/jbt.22605
Yarmohammadi F, Hayes AW, Karimi G (2021a) Protective effects of curcumin on chemical and drug-induced cardiotoxicity: a review. Naunyn Schmiedebergs Arch Pharmacol 394:1341–1353. https://doi.org/10.1007/s00210-021-02072-8
Yarmohammadi F, Karbasforooshan H, Hayes AW, Karimi G (2021b) Inflammation suppression in doxorubicin-induced cardiotoxicity: natural compounds as therapeutic options. Naunyn Schmiedebergs Arch Pharmacol 394:2003–2011. https://doi.org/10.1007/s00210-021-02132-z
Yarmohammadi F, Hesari M, Shackebaei D (2023) The role of mTOR in doxorubicin-altered cardiac metabolism: a promising therapeutic target of natural compounds. Cardiovasc Toxicol. https://doi.org/10.1007/s12012-023-09820-7
Yarmohammadi F, Hesari M, Shackebaei D (2024) The role of mTOR in doxorubicin-altered cardiac metabolism: a promising therapeutic target of natural compounds. Cardiovasc Toxicol 24:146–157
Yildirim C, Cangi S, Orkmez M et al (2023) Sinapic acid attenuated cisplatin-induced cardiotoxicity by inhibiting oxidative stress and inflammation with GPX4-mediated NF-kB modulation. Cardiovasc Toxicol 23:10–22. https://doi.org/10.1007/s12012-022-09773-3
Ying-Rui M, Bu-Fan B, Deng L et al (2023) Targeting the stimulator of interferon genes (STING) in breast cancer. Front Pharmacol 14:1199152. https://doi.org/10.3389/fphar.2023.1199152
Yüce A, Ateşşahin A, Ceribaşi AO, Aksakal M (2007) Ellagic acid prevents cisplatin-induced oxidative stress in liver and heart tissue of rats. Basic Clin Pharmacol Toxicol 101:345–349. https://doi.org/10.1111/j.1742-7843.2007.00129.x
Yum S, Li M, Fang Y, Chen ZJ (2021) TBK1 recruitment to STING activates both IRF3 and NF-κB that mediate immune defense against tumors and viral infections. Proc Natl Acad Sci U S A 118. https://doi.org/10.1073/pnas.2100225118
Zalat Z, Kohaf N, Alm El-Din M et al (2021) Silymarin: a promising cardioprotective agent. Azhar Int J Pharm Med Sci 1:13–21
Zeng Y, **ong Y, Yang T et al (2022) Icariin and its metabolites as potential protective phytochemicals against cardiovascular disease: from effects to molecular mechanisms. Biomed Pharmacother 147:112642. https://doi.org/10.1016/j.biopha.2022.112642
Zhang Q, Wu L (2022) In vitro and in vivo cardioprotective effects of curcumin against doxorubicin-induced cardiotoxicity: a systematic review. J Oncol 2022:7277562. https://doi.org/10.1155/2022/7277562
Zhang D, Lai W, Liu X et al (2021a) The safety of morphine in patients with acute heart failure: a systematic review and meta-analysis. Clin Cardiol 44:1216–1224
Zhang J, Zheng X, Wang P et al (2021b) Role of apoptosis repressor with caspase recruitment domain (ARC) in cell death and cardiovascular disease. Apoptosis 26:24–37
Zhang L-X, Li C-X, Kakar MU et al (2021c) Resveratrol (RV): a pharmacological review and call for further research. Biomed Pharmacother 143:112164. https://doi.org/10.1016/j.biopha.2021.112164
Zhao L, **ng C, Sun W et al (2018) Lactobacillus supplementation prevents cisplatin-induced cardiotoxicity possibly by inflammation inhibition. Cancer Chemother Pharmacol 82:999–1008. https://doi.org/10.1007/s00280-018-3691-8
Acknowledgements
Authors are grateful to the Kermanshah University of Medical Sciences, Health Technology Institute, Medical Biology Research Center, Kermanshah, Iran for financial support.
Funding
This work was supported by the Kermanshah University of Medical Sciences Office of Vice Chancellor for Research, Kermanshah, Iran (Grant numbers: IR.KUMS.REC.1403.021).
Author information
Authors and Affiliations
Contributions
F.Y. conceived and designed the research. M.H., P.M., and M.M. conducted screening. M.H., P.M., and M.M. extracted data. F.Y. and D.SH. wrote the manuscript. All authors read and approved the manuscript. The authors confirm that no paper mill and artificial intelligence was used.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hesari, M., Mohammadi, P., Moradi, M. et al. Molecular mechanisms involved in therapeutic effects of natural compounds against cisplatin-induced cardiotoxicity: a review. Naunyn-Schmiedeberg's Arch Pharmacol (2024). https://doi.org/10.1007/s00210-024-03207-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00210-024-03207-3