Abstract
Sulforaphane has been investigated in human pathologies and preclinical models of airway diseases. To provide further mechanistic insights, we explored L-sulforaphane (LSF) in the ovalbumin (OVA)-induced chronic allergic airways murine model, with key hallmarks of asthma. Histological analysis indicated that LSF prevented or reversed OVA-induced epithelial thickening, collagen deposition, goblet cell metaplasia, and inflammation. Well-known antioxidant and anti-inflammatory mechanisms contribute to the beneficial effects of LSF. Fourier transform infrared microspectroscopy revealed altered composition of macromolecules, following OVA sensitization, which were restored by LSF. RNA sequencing in human peripheral blood mononuclear cells highlighted the anti-inflammatory signature of LSF. Findings indicated that LSF may alter gene expression via an epigenetic mechanism which involves regulation of protein acetylation status. LSF resulted in histone and α-tubulin hyperacetylation in vivo, and cellular and enzymatic assays indicated decreased expression and modest histone deacetylase (HDAC) inhibition activity, in comparison with the well-known pan-HDAC inhibitor suberoylanilide hydroxamic acid (SAHA). Molecular modeling confirmed interaction of LSF and LSF metabolites with the catalytic domain of metal-dependent HDAC enzymes. More generally, this study confirmed known mechanisms and identified potential epigenetic pathways accounting for the protective effects and provide support for the potential clinical utility of LSF in allergic airways disease.







Similar content being viewed by others
Data and materials availability
RNA sequencing data are available from GEO under the accession GSE160353. To review GEO accession GSE160353 while it remains in private status, go the following address and enter the access token: otmlcuoillwhjcd. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160353
References
Zhang Y et al (1992) A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc Natl Acad Sci USA 89(6):2399–2403
Prochaska HJ, Santamaria AB, Talalay P (1992) Rapid detection of inducers of enzymes that protect against carcinogens. Proc Natl Acad Sci USA 89(6):2394–2398
Fahey JW et al (2015) Sulforaphane bioavailability from glucoraphanin-rich broccoli: control by active endogenous myrosinase. PLoS ONE 10(11):e0140963–e0140963
Angelino D et al (2015) Myrosinase-dependent and –independent formation and control of isothiocyanate products of glucosinolate hydrolysis. Front Plant Sci. https://doi.org/10.3389/fpls.2015.00831
Matusheski NV, Jeffery EH (2001) Comparison of the bioactivity of two glucoraphanin hydrolysis products found in broccoli, sulforaphane and sulforaphane nitrile. J Agric Food Chem 49(12):5743–5749
Tortorella SM et al (2015) Dietary sulforaphane in cancer chemoprevention: the role of epigenetic regulation and HDAC inhibition. Antioxid Redox Signal 22(16):1382–1424
Vanduchova A, Anzenbacher P, Anzenbacherova E (2018) Isothiocyanate from broccoli, sulforaphane, and its properties. J Med Food 22(2):121–126
Amjad AI et al (2015) Broccoli-derived sulforaphane and chemoprevention of prostate cancer: from bench to bedside. Current Pharmacology Reports 1(6):382–390
Zimmerman AW et al (2021) Randomized controlled trial of sulforaphane and metabolite discovery in children with Autism Spectrum Disorder. Molecular Autism 12(1):38
Sun Y et al (2020) Protective effects of sulforaphane on type 2 diabetes-induced cardiomyopathy via AMPK-mediated activation of lipid metabolic pathways and NRF2 function. Metabol Clin Experim 102:154002
Singh K et al (2014) Sulforaphane treatment of autism spectrum disorder (ASD). Proc Natl Acad Sci USA 111(43):15550–15555
Brown RH et al (2015) Sulforaphane improves the bronchoprotective response in asthmatics through Nrf2-mediated gene pathways. Respir Res 16(1):106
Jiao Z et al (2017) Sulforaphane increases Nrf2 expression and protects alveolar epithelial cells against injury caused by cigarette smoke extract. Mol Med Rep 16(2):1241–1247
Cho H-Y et al (2019) Sulforaphane enriched transcriptome of lung mitochondrial energy metabolism and provided pulmonary injury protection via Nrf2 in mice. Toxicol Appl Pharmacol 364:29–44
Sudini K et al (2016) A randomized controlled trial of the effect of broccoli sprouts on antioxidant gene expression and airway inflammation in asthmatics. J Allergy Clin Immunol Practice. 4(5):932–940
An SS et al (2016) An inflammation-independent contraction mechanophenotype of airway smooth muscle in asthma. J Allergy Clin Immunol 138(1):294-297.e4
Heber D et al (2014) Sulforaphane-rich broccoli sprout extract attenuates nasal allergic response to diesel exhaust particles. Food Funct 5(1):35–41
Mazarakis N et al (2020) The potential use of l-sulforaphane for the treatment of chronic inflammatory diseases: A review of the clinical evidence. Clin Nutr 39(3):664–675
Houghton CA (2019) Sulforaphane: its “coming of age” as a clinically relevant nutraceutical in the prevention and treatment of chronic disease. Oxid Med Cell Longev 2019:2716870–2716870
Fahey JW, Kensler TW (2021) The challenges of designing and implementing clinical trials with broccoli sprouts… and turning evidence into public health action. Front Nutr 8:183
Kensler TW et al (2012) Modulation of the metabolism of airborne pollutants by glucoraphanin-rich and sulforaphane-rich broccoli sprout beverages in Qidong, China. Carcinogenesis 33(1):101–107
Egner PA et al (2014) Rapid and sustainable detoxication of airborne pollutants by broccoli sprout beverage: results of a randomized clinical trial in China. Cancer Prevent Res (Philadelphia Pa). 7(8):813–823
De Rooij M, Jan NM, Commandeur Nico PE (1998) Vermeulen BEN Mercapturic acids as biomarkers of exposure to electrophilic chemicals:applications to environmental and industrial chemicals. Biomarkers. 3(4–5):239–303
Keum Y-S (2012) Regulation of Nrf2-mediated phase II detoxification and anti-oxidant genes. Biomol Therapeut 20(2):144–151
Al-Harbi NO et al (2019) Sulforaphane treatment reverses corticosteroid resistance in a mixed granulocytic mouse model of asthma by upregulation of antioxidants and attenuation of Th17 immune responses in the airways. Eur J Pharmacol 855:276–284
Abdull Razis AF, Iori R (2011) Ioannides C The natural chemopreventive phytochemical R-sulforaphane is a far more potent inducer of the carcinogen-detoxifying enzyme systems in rat liver and lung than the S-isomer. Int J Cancer 128(12):2775–2782
Abdull Razis AF et al (2011) Induction of epoxide hydrolase and glucuronosyl transferase by isothiocyanates and intact glucosinolates in precision-cut rat liver slices: importance of side-chain substituent and chirality. Arch Toxicol 85(8):919–927
Srovnalova A et al (2015) Effects of sulforaphane and its S- and R-enantiomers on the expression and activities of human drug-metabolizing cytochromes P450. J Funct Foods 14:487–501
Mazarakis N et al (2021) Examination of novel immunomodulatory effects of L-sulforaphane. Nutrients. 13(2):6002
Royce SG, Patel KP, Samuel CS (2014) Characterization of a novel model incorporating airway epithelial damage and related fibrosis to the pathogenesis of asthma. Lab Invest 94(12):1326–1339
Casaro M et al (2019) OVA-induced allergic airway inflammation mouse model, in Pre-Clinical Models: Techniques and Protocols., P.C. Guest, Editor. Springer New York: New York, NY. p. 297–301
Kim DI, Song M-K, Lee K (2019) Comparison of asthma phenotypes in OVA-induced mice challenged via inhaled and intranasal routes. BMC Pulm Med 19(1):241
Park JH et al (2012) Sulforaphane inhibits the Th2 immune response in ovalbumin-induced asthma. BMB Rep 45(5):311–316
Wu W et al (2019) Sulforaphane has a therapeutic effect in an atopic dermatitis murine model and activates the Nrf2/HO1 axis. Mol Med Rep 20(2):1761–1771
Yan B et al (2017) Sulforaphane prevents bleomycin-induced pulmonary fibrosis in mice by inhibiting oxidative stress via nuclear factor erythroid 2-related factor-2 activation. Mol Med Rep 15(6):4005–4014
Kim H et al (2022) A metabolomics approach to sulforaphane efficacy in secondhand smoking-induced pulmonary damage in mice. Metabolites 12(6):518
Melgert BN et al (2005) Female mice are more susceptible to the development of allergic airway inflammation than male mice. Clin Exp Allergy 35(11):1496–1503
Hayashi T et al (2003) Less sensitivity for late airway inflammation in males than females in BALB/c mice. Scand J Immunol 57(6):562–567
Royce SG et al (2011) Protective effects of valproic acid against airway hyperresponsiveness and airway remodeling in a mouse model of allergic airways disease. Epigenetics 6(12):1463–1470
Royce SG et al (2009) Relaxin reverses airway remodeling and airway dysfunction in allergic airways disease. Endocrinology 150(6):2692–2699
Mazarakis N et al (2020) Investigation of molecular mechanisms of experimental compounds in murine models of chronic allergic airways disease using synchrotron Fourier-transform infrared microspectroscopy. Sci Rep 10(1):11713
Khurana I et al (2021) SAHA attenuates Takotsubo-like myocardial injury by targeting an epigenetic Ac/Dc axis. Signal Transduct Target Ther 6(1):159
Donovan C et al (2013) Differential effects of allergen challenge on large and small airway reactivity in mice. PLoS ONE 8(9):e74101
Donovan C et al (2015) Lipopolysaccharide does not alter small airway reactivity in mouse lung slices. PLoS ONE 10(3):e0122069
Bray NL et al (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34(5):525–527
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550
Rafehi H et al (2014) Vascular histone deacetylation by pharmacological HDAC inhibition. Genome Res 24(8):1271–1284
Mootha VK et al (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34(3):267–273
Kaspi A, Ziemann M (2020) mitch: multi-contrast pathway enrichment for multi-omics and single-cell profiling data. BMC Genom 21(1):447
Wickham HD (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York
Ververis K, Karagiannis TC (2012) An atlas of histone deacetylase expression in breast cancer: fluorescence methodology for comparative semi-quantitative analysis. Am J Transl Res 4(1):24–43
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Somoza JR et al (2004) Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 12(7):1325–1334
Watson PJ et al (2012) Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 481(7381):335–340
Millard CJ et al (2013) Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol Cell 51(1):57–67
Lauffer BE et al (2013) Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J Biol Chem 288(37):26926–26943
Hai Y, Christianson DW (2016) Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol 12(9):741–747
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815
Abraham MJ et al (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25
Kim S et al (2021) PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res 49(D1):D1388-d1395
O’Boyle NM et al (2011) Open Babel: An open chemical toolbox. J Cheminform 3:33
Morris GM et al (2009) AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Bolden JE, Peart MJ, Johnstone RW (2006) Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discovery 5(9):769–784
Royce SG, Le Saux CJ (2014) Role of caveolin-1 in asthma and chronic inflammatory respiratory diseases. Expert Rev Respir Med 8(3):339–347
Gosens R et al (2009) Caveolae and Caveolins in the Respiratory System. Curr Mol Med 8:741–753
Williams TM, Lisanti MP (2004) The Caveolin genes: from cell biology to medicine. Ann Med 36(8):584–595
Bains SN et al (2012) Loss of caveolin-1 from bronchial epithelial cells and monocytes in human subjects with asthma. Allergy 67(12):1601–1604
Chen CM et al (2011) Downregulation of caveolin-1 in a murine model of acute allergic airway disease. Pediatr Neonatol 52(1):5–10
Hackett TL et al (2013) Caveolin-1 controls airway epithelial barrier function. Implications for asthma. Am J Respir Cell Mol Biol 49(4):662–71
Aravamudan B et al (2012) Caveolin-1 knockout mice exhibit airway hyperreactivity. Am J Physiol Lung Cell Mol Physiol 303(8):L669–L681
Gabehart KE et al (2013) Airway hyperresponsiveness is associated with airway remodeling but not inflammation in aging Cav1-/- mice. Respir Res 14(1):110–110
Wardyn JD, Ponsford AH, Sanderson CM (2015) Dissecting molecular cross-talk between Nrf2 and NF-κB response pathways. Biochem Soc Trans 43(4):621–626
Sun X et al (2020) NRF2 preserves genomic integrity by facilitating ATR activation and G2 cell cycle arrest. Nucleic Acids Res 48:9109–9123
Peng D et al (2019) NRF2 antioxidant response protects against acidic bile salts-induced oxidative stress and DNA damage in esophageal cells. Cancer Lett 458:46–55
Gozzelino R, Jeney V, Soares MP (2010) Mechanisms of cell protection by heme oxygenase-1. Annu Rev Pharmacol Toxicol 50(1):323–354
Kitada O et al (2001) Heme oxygenase-1 (HO-1) protein induction in a mouse model of asthma. Clin Exp Allergy 31(9):1470–1477
Dokmanovic M, Clarke C, Marks PA (2007) Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res 5(10):981
Starrett W, Blake DJ (2011) Sulforaphane inhibits de novo synthesis of IL-8 and MCP-1 in human epithelial cells generated by cigarette smoke extract. J Immunotoxicol 8(2):150–158
Elliot JG et al (2019) Fatty airways: implications for obstructive disease. Eur Respir J 54(6):1900857
Higami Y et al (2016) Increased epicardial adipose tissue is associated with the airway dominant phenotype of chronic obstructive pulmonary disease. PLoS ONE 11(2):e0148794
Adcock IM et al (2007) Epigenetic regulation of airway inflammation. Curr Opin Immunol 19(6):694–700
Royce SG, Karagiannis TC (2012) Histone deacetylases and their role in asthma. J Asthma 49(2):121–128
Ito K et al (2002) Expression and activity of histone deacetylases in human asthmatic airways. Am J Respir Crit Care Med 166(3):392–396
Barnes PJ (2010) Mechanisms and resistance in glucocorticoid control of inflammation. J Steroid Biochem Mol Biol 120(2–3):76–85
Adcock IM (2007) HDAC inhibitors as anti-inflammatory agents. Br J Pharmacol 150(7):829–831
Xu WS, Parmigiani RB, Marks PA (2007) Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene 26(37):5541–5552
Karagiannis TC, El-Osta A (2007) Will broad-spectrum histone deacetylase inhibitors be superseded by more specific compounds? Leukemia 21(1):61–65
Lunke S et al (2021) Epigenetic evidence of an Ac/Dc axis by VPA and SAHA. Clin Epigenetics 13(1):58
Pirola L et al (2011) Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Res 21(10):1601–1615
Royce SG et al (2012) Effects of the histone deacetylase inhibitor, trichostatin A, in a chronic allergic airways disease model in mice. Arch Immunol Ther Exp (Warsz) 60(4):295–306
Choi JH et al (2005) Trichostatin A attenuates airway inflammation in mouse asthma model. Clin Exp Allergy 35(1):89–96
Ren Y et al (2016) Therapeutic effects of histone deacetylase inhibitors in a murine asthma model. Inflamm Res 65(12):995–1008
Ho E, Clarke JD, Dashwood RH (2009) Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J Nutr 139(12):2393–2396
Myzak MC et al (2007) Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 232(2):227–234
Myzak MC, Ho E, Dashwood RH (2006) Dietary agents as histone deacetylase inhibitors. Mol Carcinog 45(6):443–446
Li ML et al (2020) HDAC8 inhibitor attenuates airway responses to antigen stimulus through synchronously suppressing galectin-3 expression and reducing macrophage-2 polarization. Respir Res 21(1):62
Hubbert C et al (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417(6887):455–458
Nian H et al (2009) Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds. Environ Mol Mutagen 50(3):213–221
Dashwood RH, Myzak MC, Ho E (2006) Dietary HDAC inhibitors: time to rethink weak ligands in cancer chemoprevention? Carcinogenesis 27(2):344–349
Myzak MC, Dashwood RH (2006) Histone deacetylases as targets for dietary cancer preventive agents: lessons learned with butyrate, diallyl disulfide, and sulforaphane. Curr Drug Targets 7(4):443–452
Meng XM et al (2010) Smad2 protects against TGF-beta/Smad3-mediated renal fibrosis. J Am Soc Nephrol 21(9):1477–1487
Zhang L et al (2015) Smad2 protects against TGF-β1/Smad3-mediated collagen synthesis in human hepatic stellate cells during hepatic fibrosis. Mol Cell Biochem 400(1):17–28
Fix C et al (2019) Effects of the isothiocyanate sulforaphane on TGF-β1-induced rat cardiac fibroblast activation and extracellular matrix interactions. J Cell Physiol 234(8):13931–13941
Sun C, Li S, Li D (2016) Sulforaphane mitigates muscle fibrosis in mdx mice via Nrf2-mediated inhibition of TGF-β/Smad signaling. J Appl Physiol 120(4):377–390
Javaheri B et al (2017) Stable sulforaphane protects against gait anomalies and modifies bone microarchitecture in the spontaneous STR/Ort model of osteoarthritis. Bone 103:308–317
Simões BM et al (2015) Abstract 2319: Sulforadex targets breast cancer stem-like cells in patient-derived cells and xenograft tumors. Cancer Res 75(15_Supplement): 2319–2319
Acknowledgements
We acknowledge the intellectual and financial support from McCord Research (Iowa, USA). The authors would like to acknowledge the use of the facilities provided by Monash University (Clayton, VIC, Australia) and the Murdoch Children’s Research Institute (Parkville, VIC, Australia) for their care and husbandry of the mice. The Gomori’s aldehyde-fuchsin and silver impregnation stains were performed by Ms Laura Leone at the Melbourne University Histology Platform (University of Melbourne, School of Biomedical Sciences, Parkville, VIC, Australia). The authors would like to acknowledge the use of the facilities provided by Monash Micro Imaging (MMI) at the Alfred Research Alliance (ARA, Melbourne, VIC, Australia) and, particularly, the expert assistance from Drs Stephen Cody and Iśka Carmichael. FPA-FTIR imaging measurements were undertaken at the IRM beamline at the Australian Synchrotron, part of ANSTO (Clayton, VIC, Australia). Various figures in this manuscript were created with BioRender.com. We thank the National Computing Infrastructure (NCI), and the Pawsey Supercomputing Centre in Australia (funded by the Australian Government). Further, we thank the Spartan High Performance Computing service (University of Melbourne), and the Partnership for Advanced Computing in Europe (PRACE) for awarding the access to Piz Daint, hosted at the Swiss National Supercomputing Centre (CSCS), Switzerland. We acknowledge our use of the gene set enrichment analysis, GSEA software, and Molecular Signature Database (MSigDB) (Subramanian, Tamayo, et al. (2005), PNAS 102, 15545-15550, http://www.broad.mit.edu/gsea/).
Funding
We would like to acknowledge intellectual and financial support by McCord Research (Iowa, USA). AEO is supported by an National Health and Medical Research Council (NHMRC) Senior Research Fellowship (1154650). PVL is supported by an NHMRC Career Development Fellowship (1146198). CD is supported by an NHMRC Early Career Postdoctoral Fellowship (1120152). JJL is supported by an Australian Government Research Training Program Scholarship.
Author information
Authors and Affiliations
Contributions
SGR, JEB, AH, PVL, CSS, KJS, MLKT, AE, and TCK conceptualized the ideas and overarching aims. JEB, AH, PVL, CSS, KJS, MJT, JV, MLKT, AE, and TCK were involved in supervision. KV, CD, AH, IK, JJL, SM, NM, EP, MJT, JV, and MZ participated in the development and design of the methodology. KV, CD, IK, SM, NM, MJT, JV, and MZ conducted the research and investigation process. RCB, AH, JJL, EP, YYS, and MZ were responsible for visualization and KV, EP, SGR, PVL, and TCK were involved in production of the first draft of the manuscript.
Corresponding author
Ethics declarations
Competing interests
Epigenomic Medicine Program (TCK) was supported financially by McCord Research (Iowa, USA), which has a financial interest in dietary compounds including sulforaphane. The remaining co-authors declare that they have no direct financial relation with the commercial identities mentioned in this manuscript that might lead to a conflict of interest.
Ethics approval
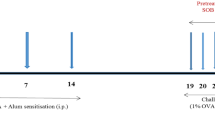
This study was performed in line with the principles of the Declaration of Helsinki. Six-week-old female BALB/c mice were obtained from Walter and Eliza Hall Institute Bioservices (prevention model; Parkville, VIC, Australia) and Monash Animal Services (reversal model; Clayton, VIC, Australia). The prevention model experimental protocol was approved by the Murdoch Children’s Research Institute Animal Ethics Committee (approval no. A597). The reversal model experimental protocol was approved by the Monash University Animal Ethics Committee (MARP/2012/085). All experimental procedures followed the Australian guidelines for the care and use of laboratory animals for scientific purposes. Human peripheral blood mononuclear cells (PBMC) were fractionated using Ficoll Paque (GE Healthcare, Wauwatosa, Wisconsin, USA) from blood samples (healthy participants; n = 4) obtained from the Australian Red Cross Blood Bank (Melbourne, VIC, Australia) under ethics project (#304/12) approved by the Alfred Hospital Ethics Committee (Alfred Health, Melbourne, VIC, Australia). Human epithelial lung A549 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Royce, S.G., Licciardi, P.V., Beh, R.C. et al. Sulforaphane prevents and reverses allergic airways disease in mice via anti-inflammatory, antioxidant, and epigenetic mechanisms. Cell. Mol. Life Sci. 79, 579 (2022). https://doi.org/10.1007/s00018-022-04609-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04609-3