Abstract
Background
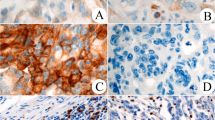
Elevated tumor-infiltrating lymphocytes (TILs) within the tumor microenvironment is a known positive prognostic factor in colorectal cancer (CRC). We hypothesized that since cytotoxic T cells release cytolytic proteins such as perforin (PRF1) and pro-apoptotic granzymes (GZMA) to attack cancer cells, a cytolytic activity score (CYT) would be a useful tool to assess anticancer immunity.
Methods
Genomic expression data were obtained from 456 patients from The Cancer Genome Atlas (TCGA). CYT was defined by GZMA and PRF1 expression, and CIBERSORT was used to evaluate intratumoral immune cell composition.
Results
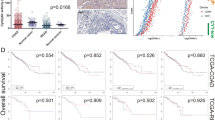
High CYT was associated with high microsatellite instability (MSI-H), as well as high levels of activated memory CD4+T cells, gamma-delta T cells, and M1 macrophages. CYT-high CRC patients had improved overall survival (p = 0.019) and disease-free survival (p = 0.016) compared with CYT-low CRC patients, especially in TIL-positive tumors. Multivariate analysis demonstrated that CYT- high associates with improved survival independently after controlling for age, lymphovascular invasion, colonic location, microsatellite instability, and TIL positivity. The levels of immune checkpoint molecules (ICMs)—programmed death-1 (PD-1), programmed death-ligand 1 (PD-L1), cytotoxic T-lymphocyte-associated protein 4 (CTLA4), lymphocyte-activation gene 3 (LAG3), T cell immunoglobulin and mucin domain 3 (TIM3), and indoleamine 2,3-dioxygenase 1 (IDO1)—correlated significantly with CYT (p < 0.0001); with improved survival in CYT-high and ICM-low patients, and poorer survival in ICM-high patients.
Conclusions
High CYT within CRC is associated with improved survival, likely due to increased immunity and cytolytic activity of T cells and M1 macrophages. High CYT is also associated with high expression of ICMs; thus, further studies to elucidate the role of CYT as a predictive biomarker of the efficacy of immune checkpoint blockade are warranted.



Similar content being viewed by others
References
Li P, **ao Z, et al. A relationship to survival is seen by combining the factors of mismatch repair status, tumor location and age of onset in colorectal cancer patients. PLoS ONE 2017; 12:e0172799.
Marmol I, Sanchez-de-Diego C, et al. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18(1):197.
Chae YK, Anker JF, et al. Genomic landscape of DNA repair genes in cancer. Oncotarget 2016; 7:23312–21.
Green AR, Aleskandarany MA, et al. Clinical impact of tumor DNA repair expression and T-cell infiltration in breast cancers. Cancer Immunol Res 2017; 5:292–99.
Park JH, Powell AG, et al. Mismatch repair status in patients with primary operable colorectal cancer: associations with the local and systemic tumour environment. Br J Cancer 2016; 114:562–70.
Prall F, Huhns M. The PD-1 expressing immune phenotype of T cell exhaustion is prominent in the ‘immunoreactive’ microenvironment of colorectal carcinoma. Histopathology 2017; 71:366–74.
Salama P, Phillips M, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol 2009; 27:186–92.
Governa V, Trella E, et al. The interplay between neutrophils and CD8(+) T cells improves survival in human colorectal cancer. Clin Cancer Res 2017; 23:3847–58.
Daster S, Eppenberger-Castori S, et al. High frequency of CD8 positive lymphocyte infiltration correlates with lack of lymph node involvement in early rectal cancer. Dis Markers 2014; 2014:792183.
Berntsson J, Nodin B, et al. Prognostic impact of tumour-infiltrating B cells and plasma cells in colorectal cancer. Int J Cancer 2016; 139:1129–39.
Woodsworth DJ, Dreolini L, et al. Targeted cell-to-cell delivery of protein payloads via the granzyme-perforin pathway. Mol Ther Methods Clin Dev 2017; 7:132–45.
Balli D, Rech AJ, et al. Immune cytolytic activity stratifies molecular subsets of human pancreatic cancer. Clin Cancer Res 2017; 23:3129–38.
Rooney MS, Shukla SA, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015; 160:48–61.
Colaprico A, Silva TC, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016; 44:e71.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490:61–70.
Cancer Genome Atlas Research Network, Weinstein JN, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013; 45:1113–20.
Crowley J, Le Blan M, Jacobson J, Salmon S. Some exploratory tools for survival analysis. Lecture Notes on Statistics. In: Proceedings of the First Seattle Symposium in Biostatistics: Survival Analysis. New York: Springer; 1997. pp. 199–229.
Newman AM, Liu CL, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015; 12:453–57.
Hause RJ, Pritchard CC, et al. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016; 22:1342–50.
Ramanathan R, Olex AL, et al. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res Treat 2017; 162:191–98.
Young J, Kawaguchi T, et al. Tamoxifen sensitivity-related microRNA-342 is a useful biomarker for breast cancer survival. Oncotarget 2017; 8:99978–89.
Kawaguchi T, Yan L, et al. Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients. Sci Rep 2017; 7:15945.
Kim SY, Kawaguchi T, et al. Clinical relevance of microRNA expressions in breast cancer validated using the cancer genome atlas (TCGA). Ann Surg Oncol 2017; 24:2943–49.
Kandoth C, McLellan MD, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502:333–39.
Markl B, Wieberneit J, et al. Number of intratumoral T lymphocytes is associated with lymph node size, lymph node harvest, and outcome in node-negative colon cancer. Am J Clin Pathol 2016; 145:826–36.
Giannakis M, Mu XJ, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep 2016; 17:1206.
Peltomaki P. DNA mismatch repair gene mutations in human cancer. Environ Health Perspect 1997; 105(Suppl 4):775–780.
MacLeod MK, Kappler JW, et al. Memory CD4 T cells: generation, reactivation and re-assignment. Immunology 2010; 130:10–15.
Yan H, Zhang P, et al. Primary Tr1 cells from metastatic melanoma eliminate tumor-promoting macrophages through granzyme B- and perforin-dependent mechanisms. Tumour Biol 2017; 39:1010428317697554.
Kiladjian JJ, Visentin G, et al. Activation of cytotoxic T-cell receptor gammadelta T lymphocytes in response to specific stimulation in myelodysplastic syndromes. Haematologica 2008; 93:381–89.
Burmeister K, Quagliata L, et al. Vascular endothelial growth factor A amplification in colorectal cancer is associated with reduced M1 and M2 macrophages and diminished PD-1-expressing lymphocytes. PLoS ONE 2017; 12:e0175563.
Holder KA, Comeau EM, et al. Origins of natural killer cell memory: special creation or adaptive evolution. Immunology 2018;154:38–49.
Lee LH, Cavalcanti MS, et al. Patterns and prognostic relevance of PD-1 and PD-L1 expression in colorectal carcinoma. Mod Pathol 2016; 29:1433–42.
Pennock GK, Chow LQ. The evolving role of immune checkpoint inhibitors in cancer treatment. Oncologist 2015; 20:812–22.
Toh JW, de Souza P, et al. The potential value of immunotherapy in colorectal cancers: review of the evidence for programmed death-1 inhibitor therapy. Clin Colorectal Cancer 2016; 15:285–91.
Prado-Garcia H, Romero-Garcia S, et al. The PD-L1/PD-1 pathway promotes dysfunction, but not “exhaustion”, in tumor-responding T cells from pleural effusions in lung cancer patients. Cancer Immunol Immunother 2017; 66:765–76.
Schreiber RD, Old LJ, et al. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 2011; 331:1565–70.
Ali HR, Chlon L, et al. Patterns of immune infiltration in breast cancer and their clinical implications: a gene-expression-based retrospective study. PLoS Med 2016; 13:e1002194.
Katz SC, Bamboat ZM, et al. Regulatory T cell infiltration predicts outcome following resection of colorectal cancer liver metastases. Ann Surg Oncol 2013; 20:946–55.
Saigusa S, Toiyama Y, et al. Implication of programmed cell death ligand 1 expression in tumor recurrence and prognosis in rectal cancer with neoadjuvant chemoradiotherapy. Int J Clin Oncol 2016; 21:946–52.
Gao Q, Qiu SJ, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol 2007; 25:2586–93.
Anderson AC. Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol Res 2014; 2:393–98.
Brahmer JR, Tykodi SS, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012; 366:2455–65.
Le DT, Uram JN, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015; 372:2509–20.
Acknowledgment
Kazuaki Takabe is supported by National Institutes of Health/National Cancer Institute (NIH/NCI) Grant R01CA160688 and Susan G. Komen Investigator Initiated Research grant IIR12222224, and is also supported by Institutional Startup Grant 71-4085-01. This work was also supported by NCI Grant P30CA016056 involving the use of Roswell Park Cancer Institute’s Bioinformatics and Biostatistics Shared Resources.
Author Contributions
SN, TK, LY, and KT had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: SN, TK, and KT. Acquisition, analysis, or interpretation of data: SN, TK, LY, QQ, and XP. Drafting of the manuscript: SN, TK, LY, QQ, XP, and KT. Critical revision of the manuscript for important intellectual content: SN, TK, and KT. Statistical analysis: TK, LY, QQ, and XP. Obtained funding: KT. Administrative, technical, or material support: LY, QQ, XP. Study supervision: TK and KT.
Disclosure
Sumana Narayanan, Tsutomu Kawaguchi, Li Yan, Xuan Peng, Qianya Qi, and Kazuaki Takabe have no potential conflicts of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
Distribution of cytolytic molecules (GZMA and PFR1) throughout the cohort of CRC patients in TCGA. Gene expression was defined as log2(TPM + 1). Supplementary Fig. S2 Kaplan–Meier survival curves of (a) overall survival and (b) disease-free survival stratified by CYT-high and CYT-low in TIL-negative patients. ‘TIL negativity’ is determined by lower CD8+ T-cell composition than the median value within the TCGA CRC cohort. Supplementary Fig. S3 (a–e) Correlation between CYT and immune checkpoint molecules: (a) IDO2, (b) TIGIT, (c) A2AR, and (d) VISTA, and (e) the inhibitory checkpoint index. The immune checkpoint index was generated by taking the log-average expression in TPM of the following molecules: PD1, PD-L1, CTLA4, A2AR TIM3, IDO1, IDO2, PD-L2, TIGIT, VISTA, and VTCN1. (f–h) Correlation between CYT and Treg markers: (f) CCR4, (g) CCR5, and (h) IL2RA. Supplementary material 1 (PPTX 462 kb)
Rights and permissions
About this article
Cite this article
Narayanan, S., Kawaguchi, T., Yan, L. et al. Cytolytic Activity Score to Assess Anticancer Immunity in Colorectal Cancer. Ann Surg Oncol 25, 2323–2331 (2018). https://doi.org/10.1245/s10434-018-6506-6
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-018-6506-6