Abstract
Background
Colorectal Cancer (CRC) is the third most common cancer type and the second leading cause of cancer-related deaths worldwide. However, the existing treatment, as well as prognosis strategies for CRC patients, need to be improved in order to increase the chance of survival. Targeted therapies of CRC, as opposed to ordinary therapies, target key biological features and pathways of cancerous cells hence minimizing the subsequent damage to normal cells. MicroRNAs have been reported to play a crucial role in inhibiting and/or suppressing major pathways in various cancer types by targeting transcripts of key genes in such pathways.
Methods
The purpose of this study was to analyze in silico the differentially expressed genes from five microarray datasets of patients with CRC. Furthermore, miRNAs were investigated to inhibit cancer cell proliferation and metastasis by targeting a key gene—frizzled receptor 3 (FZD3) in the Wnt signaling pathway.
Results
The Wnt pathway receptor FZD3 is upregulated in CRC along with other pathway genes, which play a critical role in tumorigenesis. In contrast, miR-98-5p inhibits the activity of FZD3 by binding directly to the 3′UTR of its mRNA, therefore exerting a suppressor effect on colorectal tumors.
Conclusion
The study reveals miR-98-5p as a novel target of FZD3 and an inhibitor of the Wnt signaling pathway hence being a potential candidate for develo** targeted therapies against CRC.
Similar content being viewed by others
Background
In 2020, the World Health Organization (WHO) reported that cancer was one of the leading causes of death, claiming nearly ten million lives [1]. Among all types of cancer, colon and rectal cancer, also known as colorectal cancer (CRC), is the third most commonly diagnosed type worldwide, and it caused 935,000 deaths in the year 2020, making it the second deadliest type of cancer ahead of liver, stomach, and breast cancer [2]. As of now, there are three main treatment options for CRC: surgical resection, chemotherapy, immunotherapy, or any combination of these therapies. Nevertheless, the effectiveness varies from patient to patient, especially in cases of locally invasive or metastatic cancer. We must therefore understand how this disease progresses in order to identify biomarkers that can be used to identify potential therapeutic targets in order to improve the prognosis and treatment of CRC patients.
It is possible to improve treatment for CRC with targeted therapeutic agents that target unique biological features and pathways involved in tumor progression. In contrast to other therapeutics that kill both cancerous and normal cells, these work best in cancer treatment because they target the biological features of cancerous cells only [3]. Several pathways and processes in cancerous cells can be turned off by targeted therapy, including angiogenesis, proliferation, apoptosis inhibition, differentiation, the RAS pathway, the Wnt signaling pathway, the PI3K pathway, and the cell cycle pathway [4].
A number of studies shown that activation of the highly conserved Wnt pathway in a deviant manner is a driving factor in the tumorigenesis of most human cancers, with a strong emphasis on CRC [5]. This pathway controls β-catenin, a key modulator for signal transduction in CRC through phosphorylation and ubiquitin-mediated degradation. This regulation involves key scaffold proteins such as AXIN and disheveled (DVL) which disrupt the β-catenin destruction complex that is contains 3 core proteins; adenomatous polyposis coli (APC), glycogen synthase kinase 3 beta (GSK3β), and casein kinase 1 (CK1) [6]. When the destruction complex is disrupted, β-catenin will no longer be degraded hence leading to its accumulation as free β-catenin in the cytoplasm (Piawah and Venook, 2019b), which is a hallmark of CRC progression (Cheng et al. 2019) when it translocates to the nucleus [5]. This translocated beta-catenin in association with two major transcriptional factors, i.e., T cell factor (TCF) and lymphoid enhancer-binding factor (LEF), displaces their repressor molecule Groucho. The β-catenin/TCF/LEF complex, in association with other co-activators, forms a transcriptional complex that leads to the expression of target genes of Wnt, which include MYC, CCND1, AXIN2, Cyclin D1, among others [7, 8]. These target genes are mostly oncogenes [9]. Moreover, the abnormal up-regulation of the Wnt signaling pathway is facilitated by APC mutations, which is a negative regulator of this pathway [6]. These mutations mainly lead to the loss-of-function of APC hence upregulating the Wnt signaling pathway and facilitating CRC cell proliferation and enhanced anti-apoptosis abilities through overexpression of the target genes of this pathway [9].
Wnt signaling pathway is characterized as either canonical, which is β-catenin dependent or non-canonical, which is β-catenin independent. However, the initiation of signaling events in both pathways involves the binding of Wnt molecules to frizzled receptors and other related receptors like the Low-density lipoprotein Receptor-related Protein 5/6 (LRP5/6)/ROR2/RYK for signal transduction initiation [9]. Frizzled (FZD) is a family of 10 transmembrane proteins which serve as receptors of the Wnt pathway, with every FZD member having a favored Wnt ligand [10]. Various studies have indicated that excessive activation of the Wnt signaling pathway may be a result of a loss-of-function mutation in E3 ubiquitin ligases ring-finger protein 43 (RNF43), through ubiquitin-mediated degradation blockage of frizzled receptors and LRP5/6 co-receptors. Since this is a frequently detected phenomenon in CRC [8], it, therefore, indicates that signal transduction by the Wnt pathway can be influenced as levels of expression for key components of the pathway get altered [5]. Since Wnt is the most implicated pathway in CRC, disrupting the pathway signal transduction through downregulating the expression of crucial pathway components such as FZD receptors can be a therapeutic strategy for CRC.
Human frizzled homolog 3 protein (FZD3) is located on chromosome 8p21 and is expressed in skeletal muscles, pancreas, cerebellum, stomach, kidney, and among other tissues [11]. A number of studies have shown that FZD3 is up-regulated in tissues from lung squamous cell carcinomas, lymphomas, Ewing sarcomas, and myeloma, among other cancers [12, 13]. In their study, Wong and colleagues (2013) reported that FZD3 was 100% expressed in CRC spacemen, 89% in colorectal adenomas, and 75% in colorectal polyp spacemen [12]. Therefore, there is no doubt that FZD3 plays a critical role in the development and progression of CRC, making it a potential candidate for preventative interventions.
Recent studies have demonstrated that microRNAs (miRNAs) may be effective inhibitors of proliferation, growth, and metastasis of CRC cells by targeting FZD receptors and oncogenes [13,14,15]. These single-stranded noncoding RNAs bind to the 3′ untranslated region (3′ UTR) of the target gene mRNA, thereby negatively regulating its expression. This results in the cleavage of the target gene or repression of its translation, thereby inhibiting the production of the target protein [14]. The interaction of MicroRNAs with FZD mRNAs influences the expression of FZD proteins and the Wnt pathway as a result (Smith et al. 2021). Identifying miRNAs that inhibit the expression of FZD genes in different cancers is a reported therapeutic strategy for human cancer [10]. In comparison to other FZD receptors, the FZD3 receptor has received relatively little attention in human cancers, especially colorectal cancer. It is, therefore, the purpose of this study to identify a suitable miRNA target for FZD3 receptor mRNA and to demonstrate its effectiveness as an inhibitor. The finding of this study would allow the clinical evaluation of the potential of miRNA in inhibiting CRC progression and its consideration as a therapeutic strategy in CRC treatment.
Methods
Gene expression datasets retrieval
Microarray data of five gene expression projects for normal colon and rectum samples were retrieved from the Gene Expression Omnibus database (GEO), searching against query words such as colorectal cancer and CRC, on 27th September 2021 [16]. The study’s criteria for selecting samples were Homo sapiens-derived samples, excluding cell line experiments (Table 1). In the GSE41657 dataset, there were 12 normal cells, 25 carcinoma cells, and 51 dysplastic cells. Since dysplastic cells are not true carcinoma cells, we only included the normal and carcinoma cell samples in our study.
Identification of differentially expressed genes (DEGS)
We used the limma R package (Ritchie et al. 2015) and the GEO2R tool to normalize and identify differentially expressed genes in each dataset [22]. DEGs were selected in all 5 datasets at log fold change (log FC) = 1 or − 1 and P values > 0.05 as the cut-offs. Bioinfokit v2.0.1 tool in Python was used to plot volcano plots for DEGs from each dataset [23].
Functional enrichment analysis
DEGs were arranged in descending order based on their magnitude of Log FC value after obtaining DEGs. We analyzed the top 20 DEGs from each dataset for their enrichment to understand their in-depth significance. The Supplementary Data file mentions details of the parameters submission of the top 20 DEGs. Enrichr was used to annotate the top 20 DEGs from each dataset [37]. Pathways that offer potential sites for targeted therapy in CRC include among others; Wnt/β-catenin, HGF/c-MET pathway, notch, hedgehog, and EGFR-related pathways [37]. In spite of being referred to as non-coding RNA molecules, miRNAs have been shown by a number of studies to play an important role in regulating 60% of human genes post-transcriptionally and in being associated with cancer [38]. Hence, we sought to inhibit FZD3, one of the frequently up-regulated frizzled receptors of the Wnt signaling pathway in CRC, by using miR-98-5p, a rarely reported miRNA in this disease.
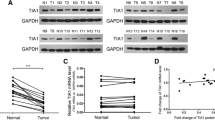
FZD3 receptor expression was determined using differential gene expression analysis on five datasets and found that the change in levels of expression between tumor and normal samples was significant enough for it to be a DEG alongside other genes. Following an analysis of gene ontology enrichment, it was found that FZD3 was significantly enriched in the Wnt signaling pathway along with many other genes within the pathway, which correlates with recent studies which implicate Wnt signaling in the development of CRC [9, 39, 40]. Further analysis of the GEDS web server datasets revealed that FZD3 expression was higher in CRCs, which validated our findings. Furthermore, we have constructed a PPI and found FZD3 receptor interacting with key WNT signaling pathway genes, including DVL, WNT1, WNT5, LRP6, and VANGL2 that were also found to be upregulated in our study data. It was also found that sFPR1 (secreted frizzled receptor protein 1), a Wnt antagonist, was downregulated in all the datasets, which was previously reported to be down-regulated by the up-regulation of FZD3 [41]. Finally, it has been reported in several studies that FZD3 expression is correlated with Wnt target genes, Cyclin D1 and c-My, which we also found to be true in our analysis [14]. Therefore, on the basis of these results and literature references, it seems reasonable to suggest that FZD3 plays a crucial role in the Wnt pathway and that its inhibition would inhibit the pathway (Fig. 8).
Recent studies have demonstrated that microRNA miR-98-5p inhibits tumor proliferation, migration and invasion by targeting the Wnt signaling pathway-related genes in various cancers including ovarian cancer [42], glioblastoma [43], gastric cancer [44] in non-small cell lung cancer [45], and pancreatic ductal adenocarcinoma [46]. The findings represent miR-98-5p as a potential target of FZD3, one of the major receptors of the Wnt pathway. Computational analysis showed that the FZD3 mRNA contained the binding sites for miR-98-5p in its 3′-UTR, which is a key feature in the miRNA post-translational gene regulation mechanism. However, miRNA prediction algorithms can barely confirm that miR-98-5p targets FZD3 directly in CRC samples. To validate the study results, luciferase reporter assays should be done to compare the behavior of a wild-type (WT) as well as the mutated (MUT) FZD3 in the 3’UTR binding site. The difference in fluorescence between FZD3-WT and FZD3-MUT will confirm that miR-98-5p is directly targeting the FZD3 gene. FZD3 and miR-98-5p could be forming an axis that inhibits Wnt signaling and CRC in general; however, the involvement of other target genes in the process cannot be ruled out. It is important that all the predicted target genes by at least two miRNA prediction algorithms are enriched in Wnt pathways by gene ontology and KEGG in order to validate the mechanism by which miR-98-5p inhibits Wnt signaling pathways. The mRNA expression levels of such genes can then be measured with miR-98-5p mimic and inhibitor, respectively.
Conclusion
In conclusion, this study demonstrated that FZD3 is upregulated in CRC along with other crucial genes of the Wnt signaling pathway. Moreover, this provides evidence that miR-98-5p may inhibit the expression of FZD3, which may lead to reduced proliferation and metastasis of colorectal cancer cells, and these findings can be used in the development of target-based therapies for CRC patients. It is essential, however, that these findings be validated by basic research in the future to determine the mechanism by which miR-98-5p regulates CRC cells, both in vivo and in vitro.
Availability of data and materials
Datasets used in this study are available in public databases as indicated in the paper citations. However, generated data and figures during this study are available on request to the corresponding author.
Abbreviations
- CRC:
-
Colorectal cancer
- miRNA:
-
MicroRNA
- FZD:
-
Frizzled receptor
- GEO:
-
Expression Omnibus database
- DEGs:
-
Differentially expressed genes
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- WNT:
-
Wingless-related integration site
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer Statistics, 2021. CA Cancer J Clin 71:7–33. https://doi.org/10.3322/caac.21654
Christodoulides N, Lami M, Malietzis G, Rasheed S, Tekkis P, Kontovounisios C (2020) Sporadic colorectal cancer in adolescents and young adults: a sco** review of a growing healthcare concern. Int J Colorectal Dis 35:1413–1421. https://doi.org/10.1007/s00384-020-03660-5
Piawah S, Venook AP (2019) Targeted therapy for colorectal cancer metastases: a review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer 125:4139–4147. https://doi.org/10.1002/cncr.32163
Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC et al (2018) Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 173:321–337.e10. https://doi.org/10.1016/j.cell.2018.03.035
Schatoff EM, Leach BI, Dow LE (2017) WNT signaling and colorectal cancer. Curr Colorectal Cancer Rep 13:101–110. https://doi.org/10.1007/s11888-017-0354-9
Novellasdemunt L, Antas P, Li VSW (2015) Targeting Wnt signaling in colorectal cancer. A review in the theme: Cell signaling: Proteins, pathways and mechanisms. Am J Physiol Cell Physiol 309:C511–21. https://doi.org/10.1152/ajpcell.00117.2015
Nie X, Liu H, Liu L, Wang YD, Chen WD (2020) Emerging roles of Wnt ligands in human colorectal cancer. Front Oncol 10. https://doi.org/10.3389/fonc.2020.01341
Cheng X, Xu X, Chen D, Zhao F, Wang W (2019) Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother 110:473–481. https://doi.org/10.1016/j.biopha.2018.11.082
Li X, Ortiz MA, Kotula L (2020) The physiological role of Wnt pathway in normal development and cancer. Exp Biol Med 245:411–426. https://doi.org/10.1177/1535370220901683
Zeng CM, Chen Z, Fu L (2018) Frizzled receptors as potential therapeutic targets in human cancers. Int J Mol Sci 19. https://doi.org/10.3390/ijms19051543
Sala CF, Formenti E, Terstappen GC, Caricasole A (2000) Identification, gene structure, and expression of human frizzled-3 (FZD3). Biochem Biophys Res Commun 273:27–34. https://doi.org/10.1006/bbrc.2000.2882
Wong SCC, He CW, Chan CML, Chan AKC, Wong HT, Cheung MT, et al (2013) Clinical significance of frizzled homolog 3 protein in colorectal cancer patients. PLoS One 8. https://doi.org/10.1371/journal.pone.0079481
Smith AJ, Sompel KM, Elango A, Tennis MA (2021) Non-coding RNA and frizzled receptors in cancer. Front Mol Biosci 8. https://doi.org/10.3389/fmolb.2021.712546
Ueno K, Hirata H, Hinoda Y, Dahiya R (2013) Frizzled homolog proteins, microRNAs and Wnt signaling in cancer. Int J Cancer 132:1731–1740. https://doi.org/10.1002/ijc.27746
Wang Q, Zhu L, Jiang Y, Xu J, Wang F, He Z (2017) miR-219-5p suppresses the proliferation and invasion of colorectal cancer cells by targeting calcyphosin. Oncol Lett 13:1319–1324. https://doi.org/10.3892/ol.2017.5570
Clough E, Barrett T (2016) The Gene Expression Omnibus database. Methods Mol Biol 1418:93–110. https://doi.org/10.1007/978-1-4939-3578-9_5
Danielsen SA, Cekaite L, Ågesen TH, Sveen A, Nesbakken A, Thiis-Evensen E, Skotheim RI, Lind GE, Lothe RA (2011) Phospholipase C Isozymes Are Deregulated in Colorectal Cancer – Insights Gained from Gene Set Enrichment Analysis of the Transcriptome. PLoS ONE 6(9):e24419. https://doi.org/10.1371/journal.pone.0024419
Del Rio M, Mollevi C, Vezzio-Vie N, Bibeau F, Ychou M, Martineau P (2013) Specific Extracellular Matrix Remodeling Signature of Colon Hepatic Metastases. PLoS ONE 8(9):e74599. https://doi.org/10.1371/journal.pone.0074599
Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA, Bujnicki JM, Menigatti M, Luz J (2007) Transcriptome Profile of Human Colorectal Adenomas Abstract. Mol Cancer Res 5(12):1263–75. https://doi.org/10.1158/1541-7786.MCR-07-0267
Gene expression profiling of colorectal normal mucosa, adenoma and adenocarcinoma tissues, NCBI GEO, GSE41657 (2012).
Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, Kirzin S (2013) Gene Expression Classification of Colon Cancer into Molecular Subtypes: Characterization Validation and Prognostic Value. PLoS Med 10(5):e1001453. https://doi.org/10.1371/journal.pmed.1001453
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Bedre R (2020) bioinfokit: Bioinformatics data analysis and visualization toolkit. Zenodo. https://doi.org/10.5281/zenodo.3747737
**e Z, Bailey A, Kuleshov M V., Clarke DJB, Evangelista JE, Jenkins SL, et al (2021) Gene Set Knowledge Discovery with Enrichr. Curr Protoc 1. https://doi.org/10.1002/cpz1.90
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4. https://doi.org/10.1186/gb-2003-4-9-r60
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J et al (2019) STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al (2003) Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M (2021) KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res 49:D545–D551. https://doi.org/10.1093/nar/gkaa970
**a M, Liu C-J, Zhang Q, Guo A-Y (2019) GEDS: A Gene Expression Display Server for mRNAs, miRNAs and Proteins. Cells 8(7):675. https://doi.org/10.3390/cells8070675
Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K et al (2013) The cancer genome atlas pan-cancer analysis project. Nat Genet 45:1113–1120. https://doi.org/10.1038/ng.2764
Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al (2016) TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 44:e71. https://doi.org/10.1093/nar/gkv1507
Robinson MD, McCarthy DJ, Smyth GK (2009) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. https://doi.org/10.1093/bioinformatics/btp616
Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, et al (2013) DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 41. https://doi.org/10.1093/nar/gkt393
Licursi V, Conte F, Fiscon G, Paci P (2019) MIENTURNET: An interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinformatics 20. https://doi.org/10.1186/s12859-019-3105-x
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4. https://doi.org/10.7554/eLife.05005
Mi B, Li Q, Li T, Liu G, Sai J (2020) High miR-31–5p expression promotes colon adenocarcinoma progression by targeting TNS1. Aging (Albany NY) 12:7480–90. https://doi.org/10.18632/AGING.103096
**e YH, Chen YX, Fang JY (2020) Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther 5. https://doi.org/10.1038/s41392-020-0116-z
Wang X, Li Y, Gong B, Zhang K, Ma Y, Li Y (2021) miR-199b-5p enhances the proliferation of medullary thymic epithelial cells via regulating Wnt signaling by targeting Fzd6. Acta Biochim Biophys Sin (Shanghai) 53:36–45. https://doi.org/10.1093/abbs/gmaa145
Li C, Nguyen V, Clark KN, Zahed T, Sharkas S, Filipp FV et al (2019) Down-regulation of FZD3 receptor suppresses growth and metastasis of human melanoma independently of canonical WNT signaling. Proc Natl Acad Sci U S A 116:4548–4557. https://doi.org/10.1073/pnas.1813802116
Patel S, Alam A, Pant R, Chattopadhyay S (2019) Wnt signaling and its significance within the tumor microenvironment: novel therapeutic insights. Front Immunol 10. https://doi.org/10.3389/fimmu.2019.02872
Cheng LC, Chao YJ, Overman MJ, Wang CY, Phan NN, Chen YL et al (2020) Increased expression of secreted frizzled related protein 1 (SFRP1) predicts ampullary adenocarcinoma recurrence. Sci Rep. https://doi.org/10.1038/s41598-020-69899-8
Wang Z, Han Y, **ng X, Jiang C, Guo H, Guan Y (2021) MiRNA-98 inhibits ovarian carcinoma cell proliferation, migration and invasion via targeting SALL4. Minerva Med 112:154–5. https://doi.org/10.23736/S0026-4806.19.06179-2
Xu X, Bao Z, Liu Y, Ji J, Liu N (2017) MicroRNA-98 Attenuates cell migration and invasion in glioma by directly targeting pre-B cell leukemia homeobox 3. Cell Mol Neurobiol 37:1359–1371. https://doi.org/10.1007/s10571-017-0466-4
Zhan P, Shu X, Chen M, Sun L, Yu L, Liu J, et al (2021) miR-98–5p inhibits gastric cancer cell stemness and chemoresistance by targeting branched-chain aminotransferases 1. Life Sci 276. https://doi.org/10.1016/j.lfs.2021.119405
Jiang F, Yu Q, Chu Y, Zhu X, Lu W, Liu Q et al (2019) MicroRNA-98-5p inhibits proliferation and metastasis in non-small cell lung cancer by targeting TGFBR1. Int J Oncol 54:128–138. https://doi.org/10.3892/ijo.2018.4610
Fu Y, Liu X, Chen Q, Liu T, Lu C, Yu J, et al (2018) Downregulated miR-98–5p promotes PDAC proliferation and metastasis by reversely regulating MAP4K4. J Exp Clin Cancer Res 37. https://doi.org/10.1186/s13046-018-0807-2
Acknowledgements
Not Applicable.
Patient consent for participation
Not applicable.
Funding
The present study did not receive any funding.
Author information
Authors and Affiliations
Contributions
Mutebi John Kenneth has designed the study, performed the experiments, and wrote the manuscript. Tushar Ahmed Shishir has performed the experiments and written the manuscript. Fahim Kabir Monjurul Haque has designed and supervised the study and written the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All the authors approved the final version of this manuscript and consent for publication.
Competing interests
Authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supplementary Data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kenneth, M.J., Shishir, T.A. & Haque, F.K.M. In silico analysis reveals mir-98-5p as a potential inhibitor of tumor cell proliferation and metastasis in colorectal cancer by targeting the fzd3 receptor of the Wnt signaling pathway. J Genet Eng Biotechnol 21, 79 (2023). https://doi.org/10.1186/s43141-023-00532-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-023-00532-7