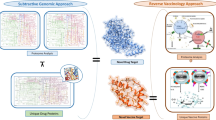
Abstract
Brucella spp. are Gram-negative, facultative intracellular bacteria that cause brucellosis in humans and various animals. The threat of brucellosis has increased, yet currently available live attenuated vaccines still have drawbacks. Therefore, subunit vaccines, produced using protein antigens and having the advantage of being safe, cost-effective and efficacious, are urgently needed. In this study, we used core proteome analysis and a compositive RV methodology to screen potential broad-spectrum antigens against 213 pathogenic strains of Brucella spp. with worldwide geographic distribution. Candidate proteins were scored according to six biological features: subcellular localization, antigen similarity, antigenicity, mature epitope density, virulence, and adhesion probability. In the RV analysis, a total 32 candidate antigens were picked out. Of these, three proteins were selected for assessment of immunogenicity and preliminary protection in a mouse model: outer membrane protein Omp19 (used as a positive control), type IV secretion system (T4SS) protein VirB8, and type I secretion system (T1SS) protein HlyD. These three antigens with a high degree of conservation could induce specific humoral and cellular immune responses. Omp19, VirB8 and HlyD could substantially reduce the organ bacterial load of B. abortus S19 in mice and provide varying degrees of protection. In this study, we demonstrated the effectiveness of this unique strategy for the screening of potential broad-spectrum antigens against Brucella. Further evaluation is needed to identify the levels of protection conferred by the vaccine antigens against wild-type pathogenic Brucella species challenge.
Similar content being viewed by others
Introduction
Brucella spp. are Gram-negative, facultative intracellular bacteria that cause brucellosis in humans and various animals [1]. The genus Brucella comprises a growing number of species (at least 12) that infect a wide variety of mammals as primary hosts [2, 3]. Brucellosis is one of the most common zoonotic diseases worldwide and has become a serious concern in recent years [4]. At present, vaccination is the most effective approach to preventing and controlling brucellosis. Veterinary live attenuated vaccines have been widely used and play an important role in the control of brucellosis epidemics [5]. However, this bacterium remains a challenging vaccine target that due to some drawbacks shown by these live attenuated vaccines, including interference with diagnostic tests, pathogenicity for humans, potential to cause abortion in pregnant animals, among others [6]. Subunit vaccines have promising applications with the advantage of being safe, cost-effective and efficacious. During the past two decades, various antigens have been extracted from Brucella, such as Omp19, Omp25, L7/L12, P39, SodC, InpB, AsnC and TF [7,8,9,10,11,12,13,14,15,16]. These available antigens have been shown to provide protection against Brucella infection by reducing the organ bacterial load in mice. Whereas such findings are highly promising, subunit vaccines using known antigens cannot provide the levels of protection conferred by live attenuated vaccines [17]. Further investigation is needed to identify novel antigens, so as to increase vaccine efficacy. B. abortus, B. melitensis, and B. suis are considered the most highly pathogenic species, causing most cases of brucellosis in humans and domestic animals throughout Central Asia, Africa, South America, and the Mediterranean region [4]. It is of great importance to select broad-spectrum antigens that can simultaneously target various Brucella pathogens with a worldwide geographic distribution.
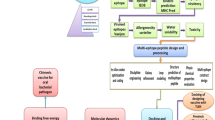
Reverse vaccinology (RV) has been proven to be a highly effective approach in which a rational vaccine design is used, with vaccine antigen prediction based on bioinformatics analysis of pathogen genomes [18, 19]. Several studies have used RV to screen potential vaccine candidates based on the protein coding genome of Brucella [ The genome of B. abortus, B. melitensis, and B. suis strains with clear geographic characteristics were selected and downloaded from the NCBI website (as of March 2019) [26]. To identify the pan-core proteome, we used an ultra-fast computational pipeline Bacterial Pan Genome Analysis (BPGA) tool with default parameters [27]. The protein FASTA files of all strains were input for orthologous cluster analysis, with an 80% sequence identity cut-off value. The core/accessory/unique proteomes were defined as coverage > 95%, 95–15%, and < 15%, respectively. Proteins from the core proteome were first aligned with host (human/mouse) protein databases using the BLASTp tool [28]. Then, non-host-homology proteins were screened and scored according to a compositive strategy assigning six biological features: (1) subcellular localization (SCL), (2) antigen similarity, (3) antigenicity, (4) mature epitope density, (5) virulence, and (6) adhesion probability. Each of the six biological features was used to divide proteins into three levels of antigen probability: high probability (individual score = 1), moderate probability (individual score = 0.5), and low probability (individual score = 0). Then, all proteins were computed and ranked based on a predicted composite score of the six individual scores for further analysis. The remaining top-ranked (1% of the core proteome) proteins were considered potential vaccine candidates. Briefly, the CELLO program was used to predict the most likely location for each protein and a subcellular localization individual score was obtained (extracellular/membrane, score = 1; periplasmic, score = 0.5; cytoplasmic, score = 0) [29]. For computation of antigen similarity, we made use of sequence similarity search programs in BLAST to identify similar sequences in the target of known protective antigens database (Protegen, exclude Brucella antigens) and obtained a similarity individual score (similarity ≥ 200, score = 1; 100 ≤ similarity < 200, score = 0.5; similarity < 100, score = 0) [ Pappas G, Akritidis N, Bosilkovski M, Tsianos E (2005) Brucellosis. N Engl J Med 352:2325–2336. https://doi.org/10.1056/NEJMra050570 Moreno E (2014) Retrospective and prospective perspectives on zoonotic brucellosis. Front Microbiol 5:213. https://doi.org/10.3389/fmicb.2014.00213 Byndloss MX, Tsolis RM (2016) Brucella spp. virulence factors and immunity. Annu Rev Anim Biosci 4:111–127. https://doi.org/10.1146/annurev-animal-021815-111326 Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV (2006) The new global map of human brucellosis. Lancet Infect Dis 6:91–99. https://doi.org/10.1016/s1473-3099(06)70382-6 Lalsiamthara J, Lee JH (2017) Development and trial of vaccines against Brucella. J Vet Sci 18:281–290. https://doi.org/10.4142/jvs.2017.18.S1.281 Dorneles EM, Sriranganathan N, Lage AP (2015) Recent advances in Brucella abortus vaccines. Vet Res 46:76. https://doi.org/10.1186/s13567-015-0199-7 Pasquevich KA, Estein SM, Garcia Samartino C, Zwerdling A, Coria LM, Barrionuevo P, Fossati CA, Giambartolomei GH, Cassataro J (2009) Immunization with recombinant Brucella species outer membrane protein Omp16 or Omp19 in adjuvant induces specific CD4+ and CD8+ T cells as well as systemic and oral protection against Brucella abortus infection. Infect Immun 77:436–445. https://doi.org/10.1128/IAI.01151-08 Goel D, Rajendran V, Ghosh PC, Bhatnagar R (2013) Cell mediated immune response after challenge in Omp25 liposome immunized mice contributes to protection against virulent Brucella abortus 544. Vaccine 31:1231–1237. https://doi.org/10.1016/j.vaccine.2012.12.043 Mallick AI, Singha H, Chaudhuri P, Nadeem A, Khan SA, Dar KA, Owais M (2007) Liposomised recombinant ribosomal L7/L12 protein protects BALB/c mice against Brucella abortus 544 infection. Vaccine 25:3692–3704. https://doi.org/10.1016/j.vaccine.2007.01.066 Gupta S, Mohan S, Somani VK, Aggarwal S, Bhatnagar R (2020) Simultaneous immunization with Omp25 and L7/L12 provides protection against Brucellosis in mice. Pathogens 9:152. https://doi.org/10.3390/pathogens9020152 Al-Mariri A, Mahmoud NH, Hammoud R (2012) Efficacy evaluation of live Escherichia coli expression Brucella P39 protein combined with CpG oligodeoxynucleotides vaccine against Brucella melitensis 16M, in BALB/c mice. Biologicals 40:140–145. https://doi.org/10.1016/j.biologicals.2012.01.002 Hop HT, Arayan LT, Huy TXN, Reyes AWB, Min W, Lee HJ, Park SJ, Chang HH, Kim S (2018) Immunization of BALB/c mice with a combination of four recombinant Brucella abortus proteins, AspC, Dps, InpB and Ndk, confers a marked protection against a virulent strain of Brucella abortus. Vaccine 36:3027–3033. https://doi.org/10.1016/j.vaccine.2018.04.019 Ghasemi A, Jeddi-Tehrani M, Mautner J, Salari MH, Zarnani AH (2015) Simultaneous immunization of mice with Omp31 and TF provides protection against Brucella melitensis infection. Vaccine 33:5532–5538. https://doi.org/10.1016/j.vaccine.2015.09.013 Wang X, An C, Yang M, Li X, Ke Y, Lei S, Xu X, Yu J, Ren H, Du X, Wang Z, Qiu Y, Liu B, Chen Z (2015) Immunization with individual proteins of the Lrp/AsnC family induces protection against Brucella melitensis 16M challenges in mice. Front Microbiol 6:1193. https://doi.org/10.3389/fmicb.2015.01193 Hou H, Liu X, Peng Q (2019) The advances in brucellosis vaccines. Vaccine 37:3981–3988. https://doi.org/10.1016/j.vaccine.2019.05.084 Masjedian Jezi F, Razavi S, Mirnejad R, Zamani K (2019) Immunogenic and protective antigens of Brucella as vaccine candidates. Comp Immunol Microbiol Infect Dis 65:29–36. https://doi.org/10.1016/j.cimid.2019.03.015 Carvalho TF, Haddad JP, Paixao TA, Santos RL (2016) Meta-analysis and advancement of brucellosis vaccinology. PLoS One 11:e0166582. https://doi.org/10.1371/journal.pone.0166582 Delany I, Rappuoli R, Seib KL (2013) Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb Perspect Med 3:a012476. https://doi.org/10.1101/cshperspect.a012476 Moxon R, Reche PA, Rappuoli R (2019) Editorial: reverse vaccinology. Front Immunol 10:2776. https://doi.org/10.3389/fimmu.2019.02776 He Y, **ang Z (2010) Bioinformatics analysis of Brucella vaccines and vaccine targets using VIOLIN. Immunome Res 6:S5. https://doi.org/10.1186/1745-7580-6-S1-S5 Gomez G, Pei J, Mwangi W, Adams LG, Rice-Ficht A, Ficht TA (2013) Immunogenic and invasive properties of Brucella melitensis 16M outer membrane protein vaccine candidates identified via a reverse vaccinology approach. PLoS One 8:e59751. https://doi.org/10.1371/journal.pone.0059751 Vishnu US, Sankarasubramanian J, Gunasekaran P, Rajendhran J (2015) Novel vaccine candidates against Brucella melitensis identified through reverse vaccinology approach. OMICS 19:722–729. https://doi.org/10.1089/omi.2015.0105 Hisham Y, Ashhab Y (2018) Identification of cross-protective potential antigens against pathogenic Brucella spp. through combining pan-genome analysis with reverse vaccinology. J Immunol Res. https://doi.org/10.1155/2018/1474517 Ong E, Wong MU, He Y (2017) Identification of new features from known bacterial protective vaccine antigens enhances rational vaccine design. Front Immunol 8:1382. https://doi.org/10.3389/fimmu.2017.01382 Dalsass M, Brozzi A, Medini D, Rappuoli R (2019) Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Front Immunol 10:113. https://doi.org/10.3389/fimmu.2019.00113 National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov Chaudhari NM, Gupta VK, Dutta C (2016) BPGA- an ultra-fast pan-genome analysis pipeline. Sci Rep 6:24373. https://doi.org/10.1038/srep24373 Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:421. https://doi.org/10.1186/1471-2105-10-421 Yu CS, Cheng CW, Su WC, Chang KC, Huang SW, Hwang JK, Lu CH (2014) CELLO2GO: a web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS One 9:e99368. https://doi.org/10.1371/journal.pone.0099368 Yang B, Sayers S, **ang Z, He Y (2011) Protegen: a web-based protective antigen database and analysis system. Nucleic Acids Res 39:D1073-1078. https://doi.org/10.1093/nar/gkq944 He Y, Racz R, Sayers S, Lin Y, Todd T, Hur J, Li X, Patel M, Zhao B, Chung M, Ostrow J, Sylora A, Dungarani P, Ulysse G, Kochhar K, Vidri B, Strait K, Jourdian GW, **ang Z (2014) Updates on the web-based VIOLIN vaccine database and analysis system. Nucleic Acids Res 42:D1124-1132. https://doi.org/10.1093/nar/gkt1133 Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform 8:4. https://doi.org/10.1186/1471-2105-8-4 Santos AR, Pereira VB, Barbosa E, Baumbach J, Pauling J, Rottger R, Turk MZ, Silva A, Miyoshi A, Azevedo V (2013) Mature Epitope Density–a strategy for target selection based on immunoinformatics and exported prokaryotic proteins. BMC Genomics 14:S4. https://doi.org/10.1186/1471-2164-14-S6-S4 Gupta A, Kapil R, Dhakan DB, Sharma VK (2014) MP3: a software tool for the prediction of pathogenic proteins in genomic and metagenomic data. PLoS One 9:e93907. https://doi.org/10.1371/journal.pone.0093907 He Y, **ang Z, Mobley HL (2010) Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J Biomed Biotechnol 2010:297505. https://doi.org/10.1155/2010/297505 Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P (2017) Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. https://doi.org/10.1093/molbev/msx148 Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. https://doi.org/10.1038/nprot.2015.053 Darzentas N (2010) Circoletto: visualizing sequence similarity with Circos. Bioinformatics 26:2620–2621. https://doi.org/10.1093/bioinformatics/btq484 Yu D, Hui Y, Zai X, Xu J, Liang L, Wang B, Yue J, Li S (2015) Comparative genomic analysis of Brucella abortus vaccine strain 104M reveals a set of candidate genes associated with its virulence attenuation. Virulence 6:745–754. https://doi.org/10.1080/21505594.2015.1038015 Wang Y, Ke Y, Wang Z, Yuan X, Qiu Y, Zhen Q, Xu J, Li T, Wang D, Huang L, Chen Z (2012) Genome sequences of three live attenuated vaccine strains of Brucella species and implications for pathogenesis and differential diagnosis. J Bacteriol 194:6012–6013. https://doi.org/10.1128/JB.01483-12 Thomas EL, Bracewell CD, Corbel MJ (1981) Characterisation of Brucella abortus strain 19 cultures. Vet Rec 108:90–93. https://doi.org/10.1136/vr.108.5.90 Qian M, Zhao T, Li R, Yang Q, Yu R, Yin Y, Zai X, Li Y, Zhang J, Xu J, Chen W (2018) Targeting the R domain of coagulase by active vaccination protects mice against lethal Staphylococcus aureus infection. Microbes Infect 21:163–169. https://doi.org/10.1016/j.micinf.2018.11.001 Lowry JE, Isaak DD, Leonhardt JA, Vernati G, Pate JC, Andrews GP (2011) Vaccination with Brucella abortus recombinant in vivo-induced antigens reduces bacterial load and promotes clearance in a mouse model for infection. PLoS One 6:e17425. https://doi.org/10.1371/journal.pone.0017425 Flower DR, Macdonald IK, Ramakrishnan K, Davies MN, Doytchinova IA (2010) Computer aided selection of candidate vaccine antigens. Immunome Res 6:S1. https://doi.org/10.1186/1745-7580-6-s2-s1 Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B (2009) Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5:580–592. https://doi.org/10.1016/j.chom.2009.05.011 Castaneda-Roldan EI, Avelino-Flores F, Dall’Agnol M, Freer E, Cedillo L, Dornand J, Giron JA (2004) Adherence of Brucella to human epithelial cells and macrophages is mediated by sialic acid residues. Cell Microbiol 6:435–445. https://doi.org/10.1111/j.1462-5822.2004.00372.x Munoz Gonzalez F, Sycz G, Alonso Paiva IM, Linke D, Zorreguieta A, Baldi PC, Ferrero MC (2019) The BtaF adhesin is necessary for full virulence during respiratory infection by Brucella suis and is a novel immunogen for nasal vaccination against Brucella infection. Front Immunol 10:1775. https://doi.org/10.3389/fimmu.2019.01775 Tibor A, Wansard V, Bielartz V, Delrue RM, Danese I, Michel P, Walravens K, Godfroid J, Letesson JJ (2002) Effect of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. Infect Immun 70:5540–5546. https://doi.org/10.1128/iai.70.10.5540-5546.2002 Risso GS, Carabajal MV, Bruno LA, Ibanez AE, Coria LM, Pasquevich KA, Lee SJ, McSorley SJ, Briones G, Cassataro J (2017) U-Omp19 from Brucella abortus is a useful adjuvant for vaccine formulations against Salmonella infection in mice. Front Immunol 8:171. https://doi.org/10.3389/fimmu.2017.00171 Pasquevich KA, Carabajal MV, Guaimas FF, Bruno L, Roset MS, Coria LM, Rey Serrantes DA, Comerci DJ, Cassataro J (2019) Omp19 enables Brucella abortus to evade the antimicrobial activity from host’s proteolytic defense system. Front Immunol 10:1436. https://doi.org/10.3389/fimmu.2019.01436 Sharifahmadian M, Nlend IU, Lecoq L, Omichinski JG, Baron C (2017) The type IV secretion system core component VirB8 interacts via the beta1-strand with VirB10. FEBS Lett 591:2491–2500. https://doi.org/10.1002/1873-3468.12770 Casu B, Mary C, Sverzhinsky A, Fouillen A, Nanci A, Baron C (2018) VirB8 homolog TraE from plasmid pKM101 forms a hexameric ring structure and interacts with the VirB6 homolog TraD. Proc Natl Acad Sci U S A 115:5950–5955. https://doi.org/10.1073/pnas.1802501115 Bielaszewska M, Aldick T, Bauwens A, Karch H (2014) Hemolysin of enterohemorrhagic Escherichia coli: structure, transport, biological activity and putative role in virulence. Int J Med Microbiol 304:521–529. https://doi.org/10.1016/j.ijmm.2014.05.005 Dorneles EMS, Teixeira-Carvalho A, Araújo MSS, Sriranganathan N, Lage AP (2015) Immune response triggered by Brucella abortus following infection or vaccination. Vaccine 33:3659–3666. https://doi.org/10.1016/j.vaccine.2015.05.057 Goolab S, Roth RL, van Heerden H, Crampton MC (2015) Analyzing the molecular mechanism of lipoprotein localization in Brucella. Front Microbiol 6:1189. https://doi.org/10.3389/fmicb.2015.01189 Lacerda TLS, Salcedo SP, Gorvel J-P (2013) Brucella T4SS: the VIP pass inside host cells. Curr Opin Microbiol 16:45–51. https://doi.org/10.1016/j.mib.2012.11.005 Coleman SA, Minnick MF (2003) Differential expression of the invasion-associated locus B (ialB) gene of Bartonella bacilliformis in response to environmental cues. Microb Pathog 34:179–186. https://doi.org/10.1016/s0882-4010(03)00005-6 Van Dyk TK, Templeton LJ, Cantera KA, Sharpe PL, Sariaslani FS (2004) Characterization of the Escherichia coli AaeAB efflux pump: a metabolic relief valve. J Bacteriol 186:7196–7204. https://doi.org/10.1128/JB.186.21.7196-7204.2004 Deng M, Misra R (1996) Examination of AsmA and its effect on the assembly of Escherichia coli outer membrane proteins. Mol Microbiol 21:605–612. https://doi.org/10.1111/j.1365-2958.1996.tb02568.x Paulley JT, Anderson ES, Roop RM 2nd (2007) Brucella abortus requires the heme transporter BhuA for maintenance of chronic infection in BALB/c mice. Infect Immun 75:5248–5254. https://doi.org/10.1128/IAI.00460-07 Fretin D, Fauconnier A, Kohler S, Halling S, Leonard S, Nijskens C, Ferooz J, Lestrate P, Delrue RM, Danese I, Vandenhaute J, Tibor A, DeBolle X, Letesson JJ (2005) The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol 7:687–698. https://doi.org/10.1111/j.1462-5822.2005.00502.x Davies MR, McIntyre L, Mutreja A, Lacey JA, Lees JA, Towers RJ, Duchene S, Smeesters PR, Frost HR, Price DJ, Holden MTG, David S, Giffard PM, Worthing KA, Seale AC, Berkley JA, Harris SR, Rivera-Hernandez T, Berking O, Cork AJ, Torres R, Lithgow T, Strugnell RA, Bergmann R, Nitsche-Schmitz P, Chhatwal GS, Bentley SD, Fraser JD, Moreland NJ, Carapetis JR et al (2019) Atlas of group A streptococcal vaccine candidates compiled using large-scale comparative genomics. Nat Genet 51:1035–1043. https://doi.org/10.1038/s41588-019-0417-8 Martirosyan A, Moreno E, Gorvel JP (2011) An evolutionary strategy for a stealthy intracellular Brucella pathogen. Immunol Rev 240:211–234. https://doi.org/10.1111/j.1600-065X.2010.00982.x Murphy EA, Sathiyaseelan J, Parent MA, Zou B, Baldwin CL (2001) Interferon-gamma is crucial for surviving a Brucella abortus infection in both resistant C57BL/6 and susceptible BALB/c mice. Immunology 103:511–518. https://doi.org/10.1046/j.1365-2567.2001.01258.x Vitry MA, De Trez C, Goriely S, Dumoutier L, Akira S, Ryffel B, Carlier Y, Letesson JJ, Muraille E (2012) Crucial role of gamma interferon-producing CD4+ Th1 cells but dispensable function of CD8+ T cell, B cell, Th2, and Th17 responses in the control of Brucella melitensis infection in mice. Infect Immun 80:4271–4280. https://doi.org/10.1128/IAI.00761-12 Zhan Y, Cheers C (1998) Control of IL-12 and IFN-gamma production in response to live or dead bacteria by TNF and other factors. J Immunol 161:1447–1453 Hop HT, Huy TXN, Reyes AWB, Arayan LT, Vu SH, Min W, Lee HJ, Kang CK, Kim DH, Tark DS, Kim S (2019) Interleukin 6 promotes Brucella abortus clearance by controlling bactericidal activity of macrophages and CD8(+) T cell differentiation. Infect Immun 87:e00431-e519. https://doi.org/10.1128/IAI.00431-19 Paul S, Peddayelachagiri BV, Nagaraj S, Konduru B, Batra HV (2018) Protective and therapeutic efficacy study of divalent fusion protein rL7/L12-Omp25 against B. abortus 544 in presence of IFNγ. Appl Microbiol Biotechnol 102:8895–8907. https://doi.org/10.1007/s00253-018-9314-9 Jankovic D, Kugler DG, Sher A (2010) IL-10 production by CD4+ effector T cells: a mechanism for self-regulation. Mucosal Immunol 3:239–246. https://doi.org/10.1038/mi.2010.8 Clapp B, Skyberg JA, Yang X, Thornburg T, Walters N, Pascual DW (2011) Protective live oral brucellosis vaccines stimulate Th1 and th17 cell responses. Infect Immun 79:4165–4174. https://doi.org/10.1128/IAI.05080-1170 Chen B, Liu B, Zhao Z, Wang G (2019) Evaluation of a DNA vaccine encoding Brucella BvrR in BALB/c mice. Mol Med Rep 19:1302–1308. https://doi.org/10.3892/mmr.2018.9735 Huang J, Pan C, Sun P, Feng E, Wu J, Zhu L, Wang HL (2020) Application of an O-linked glycosylation system in Yersinia enterocolitica serotype O:9 to generate a new candidate vaccine against Brucella abortus. Microorganisms 8:436. https://doi.org/10.3390/microorganisms8030436 Tobias L, Schurig GG, Cordes DO (1992) Comparative behaviour of Brucella abortus strains 19 and RB51 in the pregnant mouse. Res Vet Sci 53:179–183. https://doi.org/10.1016/0034-5288(92)90107-d Tobias L, Cordes DO, Schurig GG (1993) Placental pathology of the pregnant mouse inoculated with Brucella abortus strain 2308. Vet Pathol 30:119–129. https://doi.org/10.1177/030098589303000204 The Ministry of Health in China (2006) List of pathogens contagious to humans. http://www.nhc.gov.cn/qjjys/s3589/200804/f0840e5958ea40c68ba0cfda9cc138ac.shtml. Accessed 27 Jan 2006 We would like to thank Shuling Liu, Qingzhen Sun, Yujie Li, and Yue Zhang for their support in animal experiments and contribution to the reagents. This research was funded by the National Natural Science Foundation of China, grant number “31800770” and the National Major Science and Technology Project of China, grant number “2016ZX10004001”. Conceptualization, XZ, YY, JX, and WC; methodology, XZ, YY, QY and FG; formal analysis, XZ and YY; investigation, XZ, YY, QY and FG; resources, FG, RL, YL and JZ; data curation, XZ; writing—original draft preparation, XZ and YY; writing—review and editing, JX and WC; visualization, XZ; supervision, JX and WC; project administration, JX.; funding acquisition, XZ and JX. All authors read and approved the final manuscript. This study was carried out in strict accordance with the recommendations in the guidelines for the care and use of laboratory animals. All animal experiments were approved by the Bei**g Institute of Biotechnology, Bei**g, China (No. 20161101). The authors declare that they no competing interests. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Brucella spp. strains with clear genetic isolation information. Brucella proteins using compositive reverse vaccinology methodology. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data. Zai, X., Yin, Y., Guo, F. et al. Screening of potential vaccine candidates against pathogenic Brucella spp. using compositive reverse vaccinology.
Vet Res 52, 75 (2021). https://doi.org/10.1186/s13567-021-00939-5 Received: Accepted: Published: DOI: https://doi.org/10.1186/s13567-021-00939-5Materials and methods
Bioinformation and reverse vaccinology
References
Acknowledgements
Funding
Author information
Authors and Affiliations
Contributions
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Competing interests
Additional information
Publisher's Note
Supplementary Information
Additional file 1: List of 213 pathogenic
Additional file 2: The composite score for all non-host-homology
Rights and permissions
About this article
Cite this article
Keywords