Abstract
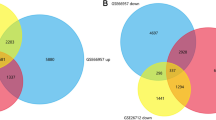
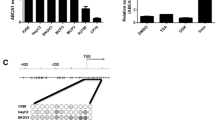
Ovarian cancer, among all gynecologic malignancies, exhibits the highest incidence and mortality rate, primarily because it is often presents with non-specific or no symptoms during its early stages. For the advancement of Ovarian Cancer Diagnosis, it is crucial to identify the potential molecular signatures that could significantly differentiate between healthy and ovarian cancerous tissues and can be used further as a diagnostic biomarker for detecting ovarian cancer. In this study, we investigated the genome-wide methylation patterns in ovarian cancer patients using Methylated DNA Immunoprecipitation (MeDIP-Seq) followed by NGS. Identified differentially methylated regions (DMRs) were further validated by targeted bisulfite sequencing for CpG site-specific methylation profiles. Furthermore, expression validation of six genes by Quantitative Reverse Transcriptase-PCR was also performed. Out of total 120 differentially methylated genes (DMGs), 68 genes were hypermethylated, and 52 were hypomethylated in their promoter region. After analysis, we identified the top 6 hub genes, namely POLR3B, PLXND1, GIGYF2, STK4, BMP2 and CRKL. Interestingly we observed Non-CpG site methylation in the case of POLR3B and CRKL which was statistically significant in discriminating ovarian cancer samples from normal controls. The most significant pathways identified were focal adhesion, the MAPK signaling pathway, and the Ras signaling pathway. Expression analysis of hypermethylated genes was correlated with the downregulation of the genes. POLR3B and GIGYF2 turned out to be the novel genes associated with the carcinogenesis of EOC. Our study demonstrated that methylation profiling through MeDIP-sequencing has effectively identified six potential hub genes and pathways that might exacerbate our understanding of underlying molecular mechanisms of ovarian carcinogenesis.
Similar content being viewed by others
Introduction
Ovarian cancer is the fifth most lethal gynecologic malignancy in women globally, mainly affecting women aged 55 to 74 years [1, 2]. It has varied heterogeneity on the molecular, histopathological, and clinical levels, and is linked with the highest fatality rates [3]. According to the American Cancer Society, a total of 19,710 new cases and 13,270 deaths have been recorded in 2023 [4]. There is a strong correlation between survival rate and stage of epithelial ovarian cancer. Early detection of ovarian cancer results in an elevated 5-year survival probability of up to 93%. However, due to the asymptomatic nature of the disease at the early stage (I/II), high recurrence rate, and lack of improved early diagnosis methods, the disease is diagnosed at an advanced stage (stage III/IV) leading to a 5-year survival rate lesser than 35% [5].
At present, the current strategies for ovarian cancer detection involve pelvic examination, transvaginal ultrasonography, and imagining techniques like MRI and PET scans. The drawback of these methods is their limited sensitivity and specificity therefore, they are combined with other serum biomarkers like CA125 and HE4 [6]. Clinically, CA125 is an FDA-approved serum biomarker routinely used for monitoring treatment and disease recurrence with a sensitivity of 50–55% and specificity of 90% [7]. Nevertheless, this sensitivity and specificity are not efficient for early-stage diagnosis and moreover CA125 is found to be elevated in benign conditions and in other non-ovarian malignancies during pregnancy. Due to the lack of early detection methods and less specificity of imaging techniques, there is an urgent need to identify a set of more valuable and reliable molecular markers and to study their role in molecular mechanisms implicated in the development and progression of ovarian cancer, which could further aid in the diagnosis of ovarian cancer.
DNA methylation is one of the most common and well-studied epigenetic modifications. Hypermethylation at the tumor suppressor gene promoters plays a key role in the onset and progression of cancer. Its high stability and occurrence in the early stage of tumorigenesis make it a promising biomarker for early detection [8]. The first direct involvement of altered DNA methylation patterns in carcinogenesis was established in 1994 by Herman et al., in cases of renal carcinoma demonstrating promoter hypermethylation as a factor responsible for the silencing of tumor suppressor gene VHL [9]. Following that, other similar investigations were undertaken, with abnormal methylation at CpG islands in the promoter region as a probable mechanism in the transcriptional suppression of tumor suppressor genes such as RASSF1a, BRCA1, CDH1, DAPK, and OPCML in a variety of cancers [10,Functional and pathway enrichment analysis An online biological information database known as the Database for Annotation, Visualisation, and Integrated Discovery (DAVID, version 6.8; http://david.ncifcrf.gov) was utilized to examine the gene ontology of DMGs [21]. This offers functional analysis according to three categories: Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analysis, Cellular Components (CC), Molecular Functions (MF), and Biological Processes (BP). P-value < 0.05 was used as the cut-off criterion for statistical significance. Search Tool for the Retrieval of Interacting Genes (STRING) (http://string-db.org/; version 11.0) is an online database for identifying the interaction between different proteins based on information fetched from sources like text mining, experiments, databases, and predictive bioinformatics data [68,69,70,71,72]. Peng et al. showed that STK4 methylation mediated downregulation consequently facilitate the progression of thyroid carcinoma by activating the Hippo signaling pathway [73]. Promoter hypermethylation of STK4 was also reported in in human sarcomas and pancreatic cancer [74, 75]. Our study presents hypermethylated CGI of POLR3B, GIGYF2, and PLXND1 with the H3k27 mark. Our preliminary results highlight targets (PLXND1, POLR3B, CRKL, GIGYF2, BMP2, and STK4) showing a negative correlation between promoter methylation gene expression. We also reported significant downregulation of these hypermethylated genes when analyzed through two GEO datasets (GSE5388 and GSE38666) and RNA-seq data (GSE212991).PPI network construction
Conclusion
This investigation showcased a comprehensive genome-wide methylation profile of epithelial ovarian cancer (EOC) and identified six hypermethylated/downregulated genes, (POLR3B, PLXND1, GIGYF2, CRKL, STK4, and BMP2) as potential diagnostic targets. The study also emphasized the potential of non-CpG sites to discriminate ovarian cancer from the disease-free normal sample. Network analysis highlighted the pathways crucial to cancer development, including the focal adhesion, Ras signaling pathway, and MAPK signaling pathway. POLR3B and GIGYF2, identified as novel hypermethylated genes, might serve as promising biomarkers for diagnosing and predicting the prognosis of ovarian cancer. These newly identified hypermethylated/downregulated genes warrant further investigations regarding their potential as therapeutic targets.
Data availability
The Medip-seq data used in this investigation is available in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE244405. Expression datasets were GSE212991, GSE54388, and GSE38666.
References
Yin F, Yi S, Wei L, Zhao B, Li J, Cai X, Liu X. Microarray-based identification of genes associated with prognosis and drug resistance in ovarian cancer. J Cell Biochem. 2019;120(4):6057–70.
Reyes HD, Devor EJ, Warrier A, Newtson AM, Mattson J, Wagner V, Gonzalez-Bosquet J. Differential DNA methylation in high-grade serous ovarian cancer (HGSOC) is associated with tumor behavior. Sci Rep. 2019;9(1):1–13.
Hatina J, Boesch M, Sopper S, Kripnerova M, Wolf D, Reimer D, Zeimet AG. Ovarian cancer stem cell heterogeneity. Stem cells heterogeneity in Cancer. Cham: Springer; 2019. pp. 201–21.
National Cancer Institute, Cancer Stat Facts. Ovarian Cancer. https://seer.cancer.gov/statfacts/html/ovary.html, 2023 (accessed May 17, 2023).
Simmons AR, Fourkala EO, Gentry-Maharaj A, Ryan A, Sutton MN, Baggerly K, Menon U. Complementary longitudinal serum biomarkers to CA125 for early detection of ovarian cancer. Cancer Prev Res. 2019;12(6):391–400.
Montagnana M, Benati M, Danese E. (2017). Circulating biomarkers in epithelial ovarian cancer diagnosis: from present to future perspective. Annals Translational Med, 5(13).
Saro**i, S., Tamir, A., Lim, H., Li, S., Zhang, S., Goy, A., … Suh, K. S. (2012).Early detection biomarkers for ovarian cancer. Journal of oncology, 2012.
Takeshima H, Yamada H, Ushijima T. Cancer epigenetics: aberrant DNA methylation in cancer diagnosis and treatment. Oncogenomics. Academic; 2019. pp. 65–76.
Hentze JL, Høgdall CK, Høgdall EV. Methylation and ovarian cancer: can DNA methylation be of diagnostic use? Mol Clin Oncol. 2019;10(3):323–30.
Li Y, Liu H, Chen H, Shao J, Su F, Zhang S, He X. DERL3 functions as a tumor suppressor in gastric cancer. Comput Biol Chem. 2020;84:107172.
Cai C, **e X, Zhou J, Fang X, Wang F, Wang M. Identification of TAF1, SAT1, and ARHGEF9 as DNA methylation biomarkers for hepatocellular carcinoma. J Cell Physiol. 2020;235(1):611–8.
Swamy SN, Premalatha CS, Pallavi VR, Gawari R. Aberrant promoter hypermethylation of RASSF1a and BRCA1 in circulating cell-free tumor DNA serves as a biomarker of ovarian carcinoma. Asian Pac J Cancer Prev. 2019;20(10):3001–5.
Barekati Z, Radpour R, Lu Q, Bitzer J, Zheng H, Toniolo P, Zhong XY. Methylation signature of lymph node metastases in breast cancer patients. BMC Cancer. 2012;12(1):244.
Ashour, N., Angulo, J. C., Andrés, G., Alelú, R., González-Corpas, A., Toledo, M.V., … Ropero, S. (2014). A DNA hypermethylation profile reveals new potential biomarkers for prostate cancer diagnosis and prognosis. The Prostate, 74(12), 1171–1182.
Liu K, Dong F, Gao H, Guo Y, Li H, Yang F, Zou C. Promoter hypermethylation of the CFTR gene as a novel diagnostic and prognostic marker of breast cancer. Cell Biol Int. 2020;44(2):603–9.
Down T. A., Rakyan V. K., Turner D. J., Flicek P., Li H., Kulesha E., Thorne N. P. A bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26(7):779–85.
Taiwo, O., Wilson, G. A., Morris, T., Seisenberger, S., Reik, W., Pearce, D., … Butcher,L. M. (2012). Methylome analysis using MeDIP-seq with low DNA concentrations. Nature protocols, 7(4), 617.
Shi Y, Gong W, Gong X, Wang P, Zhao X. Genome-wide DNA methylation analysis of breast cancer MCF-7/Taxol cells with MeDIP-Seq. PLoS ONE. 2020;15(12):e0241515. https://doi.org/10.1371/journal.pone.0241515
Lu, H., Liu, Y., Wang, J., Fu, S., Wang, L., Huang, C., … Rao, Q. (2022). Detection of ovarian cancer using plasma cell-free DNA methylomes. Clinical Epigenetics, 14(1),74.https://doi.org/10.1186/s13148-022-01285-9
Li, S., Wang, L., Zhao, Q., Wang, Z., Lu, S., Kang, Y., … Tian, J. (2020). Genome-wide analysis of cell-free DNA methylation profiling for the early diagnosis of pancreatic cancer. Frontiers in genetics, 11, 596078. https://doi.org/10.3389/fgene.2020.596078
Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44.
Yang X, Zhu S, Li L, Zhang L, **an S, Wang Y, Cheng Y. Identification of differentially expressed genes and signaling pathways in ovarian cancer by integrated bioinformatics analysis. OncoTargets Therapy. 2018;11:1457.
Rezaei-Tavirani M, Rezaei-Tavirani S, Mansouri V, Rostami-Nejad M, Rezaei-Tavirani M. Protein-protein interaction network analysis for a biomarker panel related to human esophageal adenocarcinoma. Asian Pac J cancer Prevention: APJCP. 2017;18(12):3357.
Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–9.
Mercier PL, Bachvarova M, Plante M, Gregoire J, Renaud MC, Ghani K, Bachvarov D. Characterization of DOK1, a candidate tumor suppressor gene, in epithelial ovarian cancer. Mol Oncol. 2011;5(5):438–53.
Häfner N, Steinbach D, Jansen L, Diebolder H, Dürst M, Runnebaum IB. RUNX3 and CAMK2N1 hypermethylation as prognostic marker for epithelial ovarian cancer. Int J Cancer. 2016;138(1):217–28.
Kang S, Dong SM, Park NH. Frequent promoter hypermethylation of TGFBI in epithelial ovarian cancer. Gynecol Oncol. 2010;118(1):58–63.
Ozdemir F, Altinisik J, Karateke A, Coksuer H, Buyru N. Methylation of tumor suppressor genes in ovarian cancer. Experimental Therapeutic Med. 2012;4(6):1092–6.
Kim M, Costello J. DNA methylation: an epigenetic mark of cellular memory. Exp Mol Med. 2017;49(4):e322–322. https://doi.org/10.1038/emm.2017.10
Barton CA, Hacker NF, Clark SJ, O’Brien PM. DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol. 2008;109(1):129–39.
Gloss, B. S., Patterson, K. I., Barton, C. A., Gonzalez, M., Scurry, J. P., Hacker,N. F., … Clark, S. J. (2012). Integrative genome-wide expression and promoter DNA methylation profiling identifies a potential novel panel of ovarian cancer epigenetic biomarkers. Cancer letters, 318(1), 76–85.
Tilghman RW, Parsons JT. (2008, February). Focal adhesion kinase as a regulator of cell tension in the progression of cancer. In Seminars in cancer biology (Vol. 18, No. 1, pp. 45–52). Academic Press. https://doi.org/10.1016/j.semcancer.2007.08.002
Hendrikse CSE, Theelen PMM, van der Ploeg P, Westgeest HM, Boere IA, Thijs AMJ… Piek, J. M. J. The potential of RAS/RAF/MEK/ERK(MAPK) signaling pathway inhibitors in ovarian cancer: a systematic review and meta-analysis. Gynecol Oncol. 2023;171:83–94. https://doi.org/10.1016/j.ygyno.2023.01.038
Moufarrij, S., Dandapani, M., Arthofer, E., Gomez, S., Srivastava, A., Lopez-Acevedo,M., … Chiappinelli, K. B. (2019). Epigenetic therapy for ovarian cancer: promise and progress. Clinical epigenetics, 11(1), 1–11.https://doi.org/10.1186/s13148-018-0602-0
Therachiyil L, Anand A, Azmi A, Bhat A, Korashy HM, Uddin S. Role of RAS signaling in ovarian cancer. F1000Research. 2022;11. https://doi.org/10.12688/f1000research.126337.1
Ziller MJ, Müller F, Liao J, Zhang Y, Gu H, Bock C, Meissner A. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7(12):e1002389.
Ramasamy D, Magendhra Rao D, Rajkumar AK, T., Mani S. Non-CpG methylation—a key epigenetic modification in cancer. Brief Funct Genomics. 2021;20(5):304–11.
**ao W, Liu X, Niu X, Li C, Guo Y, Tan J, Li Y. The frequency of CpG and non-CpG methylation of Notch3 gene promoter determines its expression levels in breast cancer cells. Exp Cell Res. 2020;386(2):111743.
Ishii T, Fujishiro M, Masuda M, Teramoto S, Matsuse T. A methylated oligonucleotide induced methylation of GSTP1 promoter and suppressed its expression in A549 lung adenocarcinoma cells. Cancer Lett. 2004;212(2):211–23.
Kouidou, S., Agidou, T., Kyrkou, A., Andreou, A., Katopodi, T., Georgiou, E., … Tzimagiorgis,G. (2005). Non-CpG cytosine methylation of p53 exon 5 in non-small cell lung carcinoma.Lung Cancer, 50(3), 299–307.
Patil V, Ward RL, Hesson LB. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics. 2014;9(6):823–8. https://doi.org/10.4161/epi.28741
Ramasamy D, Magendhra Rao D, Rajkumar AK, T., Mani S. Non-CpG methylation—a key epigenetic modification in cancer. Brief Funct Genomics. 2021;20(5):304–11. https://doi.org/10.1093/bfgp/elab035
Luo WM, Wang ZY, Zhang X. Identification of four differentially methylated genes as prognostic signatures for stage I lung adenocarcinoma. Cancer Cell Int. 2018;18(1):1–8. https://doi.org/10.1186/s12935-018-0547-6
Han Y, Zheng Q, Tian Y, Ji Z, Ye H. Identification of a nine-gene panel as a prognostic indicator for recurrence with muscle‐invasive bladder cancer. J Surg Oncol. 2019;119(8):1145–54.https://doi.org/10.1002/jso.25446
Zhu, L., Yao, Z., Luo, Q., Liu, Y., Zhao, W., Shao, C., … Cui, F. (2023). Low Expression of GIGYF1 Inhibits Metastasis, Proliferation, and Promotes Apoptosis and Autophagy of Gastric Cancer Cells. International Journal of Medical Sciences, 20(8), 1038–1045.https://doi.org/10.7150/ijms.82719
Yang, W., Yuan, Q., Zhang, S., Zuo, M., Li, T., Li, J., … Liu, Y. (2022). Elevated GIGYF2 expression suppresses tumor migration and enhances sensitivity to temozolomide in malignant glioma. Cancer Gene Therapy, 29(6), 750–757.https://doi.org/10.1038/s41417-021-00353-1
Vivekanadhan S, Mukhopadhyay D. (2019). Divergent roles of Plexin D1 in cancer. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 1872(1), 103–10. https://doi.org/10.1016/j.bbcan.2019.05.004
Li J, Hu K, He D, Zhou L, Wang Z, Tao Y. Prognostic value of PLXND1 and TGF-β1 coexpression and its correlation with immune infiltrates in hepatocellular carcinoma. Front Oncol. 2021;10:604131. https://doi.org/10.3389/fonc.2020.604131
Hagihara, K., Haraguchi, N., Nishimura, J., Yasueda, A., Fu**o, S., Ogino, T., …Eguchi, H. (2022). PLXND1/SEMA3E Promotes Epithelial–Mesenchymal Transition Partly via the PI3K/AKT-Signaling Pathway and Induces Heterogenity in Colorectal Cancer.Annals of Surgical Oncology, 29(12), 7435–7445. https://doi.org/10.1245/s10434-022-11945-y.
Tseng CH, Murray KD, Jou MF, Hsu SM, Cheng HJ, Huang PH. Sema3E/plexin-D1 mediated epithelial-to-mesenchymal transition in ovarian endometrioid cancer. PLoS ONE. 2011;6(4):e19396. https://doi.org/10.1371/journal.pone.0019396
Shalaby MA, Hampson L, Oliver A, Hampson I. Plexin D1: new potential biomarker for cervical cancer. J Immunoass Immunochemistry. 2012;33(3):223–33. https://doi.org/10.1080/15321819.2011.634472
Rehman M, Gurrapu S, Cagnoni G, Capparuccia L, Tamagnone L. PlexinD1 is a novel transcriptional target and effector of notch signaling in cancer cells. PLoS ONE. 2016;11(10):e0164660. https://doi.org/10.1371/journal.pone.0164660
Fu L, Dong Q, **e C, Wang Y, Li Q. CRKL protein overexpression enhances cell proliferation and invasion in pancreatic cancer. Tumor Biology. 2015;36:1015–22. https://doi.org/10.1007/s13277-014-2706-2
Zhao, T., Miao, Z., Wang, Z., Xu, Y., Wu, J., Liu, X., … Li, J. (2013). Overexpression of CRKL correlates with malignant cell proliferation in breast cancer. Tumor Biology,34, 2891–2897. https://doi.org/10.1007/s13277-013-0851-7
Wang Y, Dong QZ, Fu L, Stoecker M, Wang E, Wang EH. Overexpression of CRKL correlates with poor prognosis and cell proliferation in non-small cell lung cancer. Mol Carcinog. 2013;52(11):890–9. https://doi.org/10.1002/mc.21935
Natsume, H., Shinmura, K., Tao, H., Igarashi, H., Suzuki, M., Nagura, K., … Sugimura,H. (2012). The CRKL gene encoding an adaptor protein is amplified, overexpressed,and a possible therapeutic target in gastric cancer. Journal of translational medicine,10(1), 1–11. https://doi.org/10.1186/1479-5876-10-97.
Li, Z., Wu, X., Chen, S., Zhong, J., Qiu, X., Kpegah, J. K., … Zhou, J. (2023). Identification of CRKL as an oncogenic biomarker for prognosis and immunotherapy in melanoma, and its potential molecular mechanism. Genomics, 115(3), 110634.https://doi.org/10.1016/j.ygeno.2023.110634.
Cheng S, Guo J, Yang Q, Yang X. Crk-like adapter protein regulates CCL19/CCR7-mediated epithelial-to-mesenchymal transition via ERK signaling pathway in epithelial ovarian carcinomas. Med Oncol. 2015;32:1–7. https://doi.org/10.1007/s12032-015-0494-1
Wang J, Che YL, Li G, Liu B, Shen TM, Wang H, Linghu H. Crk and CrkL present with different expression and significance in epithelial ovarian carcinoma. Mol Carcinog. 2011;50(7):506–15. https://doi.org/10.1002/mc.20745
Li TT, Lai YW, Han X, Niu X, Zhang PX. BMP2 as a promising anticancer approach: functions and molecular mechanisms. Investig New Drugs. 2022;40(6):1322–32. https://doi.org/10.1007/s10637-022-01298-4
Bieniasz M, Oszajca K, Eusebio M, Kordiak J, Bartkowiak J, Szemraj J. The positive correlation between gene expression of the two angiogenic factors: VEGF and BMP-2 in lung cancer patients. Lung Cancer. 2009;66(3):319–26. https://doi.org/10.1016/j.lungcan.2009.02.020
Vishnubalaji R, Yue S, Alfayez M, Kassem M, Liu FF, Aldahmash A, Alajez NM. Bone morphogenetic protein 2 (BMP2) induces growth suppression and enhances chemosensitivity of human colon cancer cells. Cancer Cell Int. 2016;16(1):1–12. https://doi.org/10.1186/s12935-016-0355-9
Kang MH, Kim JS, Seo JE, Oh SC, Yoo YA. BMP2 accelerates the motility and invasiveness of gastric cancer cells via activation of the phosphatidylinositol 3-kinase (PI3K)/Akt pathway. Exp Cell Res. 2010;316(1):24–37. https://doi.org/10.1016/j.yexcr.2009.10.010
Hsu, Y. T., Gu, F., Huang, Y. W., Liu, J., Ruan, J., Huang, R. L., … Huang, T. H.M. (2013). Promoter hypomethylation of EpCAM-regulated bone morphogenetic protein gene family in recurrent endometrial cancer. Clinical Cancer Research, 19(22), 6272–6285.https://doi.org/10.1158/1078-0432.CCR-13-1734
Le Page, C., Puiffe, M. L., Meunier, L., Zietarska, M., de Ladurantaye, M., Tonin,P. N., … Mes-Masson, A. M. (2009). BMP-2 signaling in ovarian cancer and its association with poor prognosis. Journal of ovarian research, 2(1), 1–11.https://doi.org/10.1186/1757-2215-2-4
Ma Y, Ma L, Guo Q, Zhang S. Expression of bone morphogenetic protein-2 and its receptors in epithelial ovarian cancer and their influence on the prognosis of ovarian cancer patients. J Experimental Clin Cancer Res. 2010;29(1):1–6. https://doi.org/10.1186/1756-9966-29-85
Fukuda, T., Fukuda, R., Tanabe, R., Koinuma, D., Koyama, H., Hashizume, Y., … Heldin,C. H. (2020). BMP signaling is a therapeutic target in ovarian cancer. Cell Death Discovery, 6(1), 139. https://doi.org/10.1038/s41420-020-00377-w
Ready D, Yagiz K, Amin P, Yildiz Y, Funari V, Bozdag S, Cinar B. Map** the STK4/Hippo signaling network in prostate cancer cell. PLoS ONE. 2017;12(9):e0184590. https://doi.org/10.1371/journal.pone.0184590
Govorov I, Attarha S, Kovalevska L, Andersson E, Kashuba E, Mints M. STK4 protein expression pattern follows different trends in endometrioid and serous endometrial adenocarcinoma upon tumor progression. Sci Rep. 2022;12(1):22154. https://doi.org/10.1038/s41598-022-26391-9
Chao, Y., Wang, Y., Liu, X., Ma, P., Shi, Y., Gao, J., … Zhou, X. (2015). Mst1 regulates glioma cell proliferation via the AKT/mTOR signaling pathway. Journal of neuro-oncology,121, 279–288. https://doi.org/10.1007/s11060-014-1654-4
Lin, C. H., Hsu, T. I., Chiou, P. Y., Hsiao, M., Wang, W. C., Chen, Y. C., … Lu, P.J. (2020). Downregulation of STK4 promotes colon cancer invasion/migration through blocking β-catenin degradation. Molecular Oncology, 14(10), 2574–2588.https://doi.org/10.1002/1878-0261.12771
Bai ZY, Peng LS, Li RQ, Peng XC, Yang Z. (2022). STK4, a novel prognostic biomarker, correlated with immune infiltrates in clear cell renal cell carcinoma. https://doi.org/10.21203/rs.3.rs-2340697/v1
Peng, X., Ji, C., Tan, L., Lin, S., Zhu, Y., Long, M., … Li, H. (2020). Long non-coding RNA TNRC6C‐AS1 promotes methylation of STK4 to inhibit thyroid carcinoma cell apoptosis and autophagy via Hippo signalling pathway. Journal of Cellular and Molecular Medicine,24(1), 304–316.https://doi.org/10.1111/jcmm.14728
Seidel, C., Schagdarsurengin, U., Blümke, K., Würl, P., Pfeifer, G. P., Hauptmann,S., … Dammann, R. (2007). Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Molecular Carcinogenesis: Published in cooperation with the University of Texas MD Anderson Cancer Center, 46(10), 865–871.https://doi.org/10.1002/mc.20317.
Mirus JE, Zhang Y, Hollingsworth MA, Solan JL, Lampe PD, Hingorani SR. Spatiotemporal proteomic analyses during pancreas cancer progression identifies serine/threonine stress kinase 4 (STK4) as a novel candidate biomarker for early stage disease. Mol Cell Proteom. 2014;13(12):3484–96. https://doi.org/10.1074/mcp.M113.036517
Acknowledgements
The authors express their gratitude to all the participants and control groups who participated in the investigation. The authors also thank King George Medical College, Lucknow, India, for providing the ovarian samples used in this research. Additionally, the authors would like to acknowledge the Central Instrumentation Facility of MNNIT Allahabad for granting access to Real-time PCR resources. Their appreciation also extends to Redcliffe Life Sciences, New Delhi, for providing the sequencing facility.
Funding
Indian Council of Medical Research (ICMR) provided funds in this project. (Sanction Order No. 5/13/58/2015/NCD-III), New Delhi.
Author information
Authors and Affiliations
Contributions
P.G: Writing- Original draft preparation, performed the experiment, software, Visualization and data curation. S.G: gave the ovarian cancer samples, Review & Editing. M.S: designing the experiment, supervision, writing- review and editing. Before submission, the final draft of the work was reviewed and approved by all authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The MNNIT Allahabad, Institute Ethics Committee granted permission for the use of human ovarian cancer and normal tissues.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gautam, P., Gupta, S. & Sachan, M. Comprehensive DNA methylation profiling by MeDIP-NGS identifies potential genes and pathways for epithelial ovarian cancer. J Ovarian Res 17, 83 (2024). https://doi.org/10.1186/s13048-024-01395-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13048-024-01395-3