Abstract
Background
Miscarriage is a frustrating complication of pregnancy that is common among women of reproductive age. Insufficient decidualization which not only impairs embryo implantation but disturbs fetomaternal immune-tolerance, has been widely regarded as a major cause of miscarriage; however, the underlying mechanisms resulting in decidual impairment are largely unknown.
Methods
With informed consent, decidual tissue from patients with spontaneous abortion or normal pregnant women was collected to detect the expression profile of UCHL1. Human endometrial stromal cells (HESCs) were used to explore the roles of UCHL1 in decidualization and dNK modulation, as well as the mechanisms involved. C57/BL6 female mice (7–10 weeks old) were used to construct pregnancy model or artificially induced decidualization model to evaluate the effect of UCHL1 on mice decidualization and pregnancy outcome.
Results
The Ubiquitin C-terminal hydrolase L1 (UCHL1), as a deubiquitinating enzyme, was significantly downregulated in decidua from patients with miscarriage, along with impaired decidualization and decreased dNKs. Blockage of UCHL1 led to insufficient decidualization and resultant decreased expression of cytokines CXCL12, IL-15, TGF-β which were critical for generation of decidual NK cells (dNKs), whereas UCHL1 overexpression enhanced decidualization accompanied by increase in dNKs. Mechanistically, the promotion of UCHL1 on decidualization was dependent on its deubiquitinating activity, and intervention of UCHL1 inhibited the activation of JAK2/STAT3 signaling pathway, resulting in aberrant decidualization and decreased production of cytokines associated with dNKs modulation. Furthermore, we found that inhibition of UCHL1 also disrupted the decidualization in mice and eventually caused adverse pregnancy outcome.
Conclusions
UCHL1 plays significant roles in decidualization and dNKs modulation during pregnancy in both humans and mice. Its deficiency indicates a poor pregnancy outcome due to defective decidualization, making UCHL1 a potential target for the diagnosis and treatment of miscarriage.
Graphical Abstract

Similar content being viewed by others
Background
Miscarriage, also known as spontaneous abortion, is a distressing reproductive complication during early pregnancy that affects 15.3% of clinically recognized pregnancies [1], and it can be caused by various factors, such as chromosomal errors, uterine infections, and endocrine abnormalities [2]. Decidualization defect is one of the major maternal causes of miscarriage [3]; however, the underlying molecular mechanisms regulating decidualization and the pathology of miscarriage related to impaired decidualization remain largely unknown.
Decidualization refers to the transformation of endometrial stromal cells (ESCs) into specialized secretory decidual stromal cells (DSCs), which provide a ‘fertile ground’ for embryo implantation [4, 5]. Moreover, the decidualized endometrium plays crucial roles in protecting the embryo from maternal immune rejection by communicating with decidual immune cells (DICs) to establish an immune-tolerant niche at the maternal–fetal interface [6]. Decidual NK cells (dNKs) are the predominant immune population and account for approximately 70% of all DICs in early-pregnancy decidua [6], and their recruitment and education are largely dependent on chemokines and cytokines secreted by DSCs [7,8,9]. Hence, defective decidualization not only impedes embryo implantation but also disrupts the development of fetomaternal immunotolerance, ultimately leading to miscarriage. Accordingly, the molecular mechanism underlying aberrant decidualization is an urgent issue to be explored.
Ubiquitination and deubiquitination are dynamic, multifaceted post-translational modifications that play a dominant role in protein function and degradation [10]. This modification, orchestrated by numerous ubiquitin ligases or deubiquitinating enzymes, controls almost all fundamental biological processes of metazoans, including cell division, differentiation and migration [11]. Increasing evidences suggest that ubiquitinating and deubiquitinating modifications are involved in pregnancy. For example, loss of the deubiquitinating enzyme USP36 causes preimplantation embryonic lethality in mice by modulating the stability of DHX33 [12]. Cullin 1 (CUL1), a scaffold protein in cullin-based ubiquitin ligases, promotes trophoblast invasion and migration [13]. Moreover, pregnancy complications, especially miscarriage, have been reported to be related to dysregulated ubiquitination [14,15,16]. However, it is unclear whether the processes of ubiquitination and deubiquitination modulate endometrial decidualization.
Ubiquitin C-terminal hydrolase L1 (UCHL1) is a deubiquitinating enzyme that was first studied in the nervous system and is associated with synaptic function and contextual memory [17]. Aberrant function of UCHL1 has been reported to be involved in neurodegenerative disorders such as Parkinson’s disease [18] and Alzheimer’s disease [19]. Moreover, UCHL1 has been reported as a multifunctional deubiquitinating enzyme involved in cancer invasion, metastasis [20] and chemoresistance [21]. Our previous study revealed that UCHL1 negatively regulated the immunosuppressive activity of mesenchymal stem cells [22]. Additionally, within the mammalian reproductive system, UCHL1 plays an essential regulatory role in controlling mammalian oocyte development and spermatogenesis [23]. A recent finding reveals that within the uterus during early pregnancy of mice, UCHL1 is specifically expressed in decidual cells [24]; however, the roles of UCHL1 have not been explored in miscarriage related to impaired decidualization.
In the present study, we found that the expression of UCHL1 was dramatically decreased in decidua from patients suffering miscarriage, accompanied by aberrant decidualization indicated by the downregulation of decidual markers IGFBP1 and PRL [25] and decreased dNKs due to reduced production of CXCL12, IL15 and TGF-β from DSCs. Blocking UCHL1 inhibited endometrial decidualization and resulted in decreased percentage of dNKs. Conversely, UCHL1 overexpression enhanced the decidualization of ESCs and then promoted the recruitment and education of dNKs, which was dependent on its deubiquitinating activity. Activation of the JAK2/STAT3 signaling pathway, which is essential for decidualization [26], was inhibited when UCHL1 was treated with its specific inhibitor or shRNA but was enhanced in UCHL1-overexpressing ESCs. Furthermore, the enhanced decidualization and cytokine expression induced by UCHL1 overexpression were reversed by STAT3 inhibition. These results indicated that the promotion of decidualization by UCHL1 was dependent on the activation of JAK/STAT3 signaling. Moreover, we verified that UCHL1 played an important role in maintaining successful decidualization and pregnancy in mice, suggesting a conserved function of UCHL1 in pregnancy, which made it feasible to test the efficacy of medicine targeting UCHL1 on miscarriage in a mouse model. Our findings provide evidence that UCHL1 is critical for decidualization as well as the recruitment and education of dNKs and therefore represents a potential target for the diagnosis and treatment of miscarriage.
Methods
Human sample collection
This study was approved by the Medical Research Ethics Committee of the Affiliated Changshu Hospital of Nantong University, Changshu, China (ethics approval number: 2018037). Written informed consent was obtained from all subjects. Human endometrial samples and peripheral blood samples were collected from patients with clinically normal pregnancies terminated for nonmedical reasons (n = 20) and unexplained spontaneous abortions (n = 20) at the Affiliated Changshu Hospital of Nantong University, Changshu, China. Criteria for a normal pregnancy include no history of spontaneous abortion or abnormal birth, absence of abdominal pain, vaginal bleeding, fever, or any other evident discomfort after conception. Preoperative examination revealed no pathogen infection and ultrasound examination confirmed the presence of original cardiac tube pulsation without significant uterine effusion. The criteria for unexplained spontaneous abortion include the absence of fetal heart tube pulsation during two consecutive vaginal ultrasound examinations conducted one week apart in early pregnancy. Other potential causes such as trauma, fever, genital malformation, infection, or chromosome abnormality were ruled out. Clinical characteristics of patients with unexplained spontaneous abortions and clinically normal pregnancy are presented in supplementary Table 2.
Animals
C57BL/6 mice were purchased from the Shanghai Laboratory Animal Center of the Chinese Academy of Sciences (CAS, Shanghai, China). The animals were housed in a specific pathogen-free (SPF) facility at Shanghai Institutes for Biological Sciences, CAS. All animal experiments were conducted following the guidelines of the Institutional Animal Care and Use Committee of Shanghai Institutes for Biological Sciences, CAS (ethics approval number: 20200306-028), and complied with the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health.
Mouse model
Adult C57BL/6 mice (7–10 weeks old) were mated to establish pregnancy. The presence of a vaginal plug was considered day 0 of gestation. To block UCHL1 in-vivo, a working solution was prepared by adding 50 μL of LDN57444 (dissolved in DMSO at 4 mg/mL) or DMSO to 950 μL of corn oil. Subsequently, the mice were injected i.p. daily with the identical volume of working solution containing DMSO (control group) or 0.4 mg/kg LDN57444 (UCHL1 blockade group; S7135, Selleckchem, USA) from gestational day (GD) 5 to 7 (n = 6 per group). The mice were sacrificed on GD8 and the uterine tissues were collected.
The establishment of artificially induced decidualization has been previously described [27]. Briefly, C57BL/6 female mice at 7–8 weeks of age were ovariectomized (n = 6 per group). Two weeks after ovariectomy, the mice were injected with 100 ng of E2 daily for three days. After a two-day interval, the mice received daily injections of 1 mg of P4 and 6.7 ng of E2. Six hours after the third E2 and P4 injection, the right uterine horn was stimulated by 50 µL of sesame oil, while the contralateral horn remained untreated as a control. Daily injection of E2 and P4 was continued for 5 days to induce decidualization of the endometrium. The mice were sacrificed on Day 6 for tissue collection.
Isolation of DSCs and DICs
Decidual tissues were washed twice with PBS plus penicillin–streptomycin solution (P/S), and DSCs were isolated as soon as possible according to the previously described method [28]. Briefly, decidual tissues were cut into pieces and digested with 0.1% collagenase type IV (Roche, Vienna, Austria) and 0.002% DNase I (Sigma-Aldrich, Darmstadt, Germany) in DMEM/F-12 medium for 30 to 60 min at 37 °C. After digestion, the tissue pieces were filtered through sterile gauze pads (200 mesh) to remove cellular debris. The cell suspension was then centrifuged at 1500 rpm for 8 min, and the supernatant was discarded. Collected cells were resuspended in DMEM/F-12 medium, layered over different concentrations (60%, 40% and 20% from bottom top) of Percoll (GE Healthcare, Chicago, United States) and centrifuged at 2000 rpm for 20 min. The cells from the 20%/40% interface mainly consisted of DSCs, while those from the 40%/60% interface mainly consisted of decidual immunocytes (DICs). Finally, isolated DSCs were cultured in dishes with DMEM/F-12 supplemented with 10% FBS and incubated in a humidified incubator with 5% CO2 at 37 °C. Isolated DICs were cultured in RPMI-1640 medium (HyClone, Utah, United States) containing 10% FBS (Gibco, Massachusetts, United States) and 50 µg/mL penicillin/streptomycin.
pNK cell isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples collected from healthy pregnant women in the first trimester by Ficoll-Hypaque (Sigma-Aldrich) density gradient centrifugation at 800×g for 20 min. According to the manufacturer’s instructions (MiltenyiBiotec, Bergisch Gladbach, Germany), peripheral NK cells (pNKs) were obtained through negative selection using the human NK cell isolation kit. pNKs were cultured in RPMI-1640 medium (HyClone) supplemented with 10% FBS (Gibco) and 50 µg/mL penicillin/streptomycin. The purity of CD45+CD3−CD56+ pNKs was > 90%, as determined by flow cytometry assays (data not shown).
Cell coculture assay
For the transwell assay, DSCs isolated from different samples were cultured in medium as described above and treated with LDN57444 or identical volume of DMSO as indicated. The culture medium was then collected in 24-well plates. pNKs (1 * 105 cells/well) were placed in the upper compartment of the Transwell chamber inserts (5 μm aperture, Corning, Painted Post, NY, USA). After 48 h of coculture, the number of dNKs in the lower compartments was counted.
For the coculture assay, isolated DSCs or decidualized HESCs (described below) were seeded in 24-well plates and treated LDN57444 or identical volume of DMSO as control for 24 h. Then, the culture medium was discarded and pNKs were seeded into the plate to coculture with these cells for another 48 h. The suspended cells were collected for flow cytometric analysis.
Flow cytometry assays
pNKs cocultured with DSCs or decidual HESCs were collected, and the expression of CD45, CD3, CD56 and CD16 was analyzed by flow cytometry assays. Briefly, the collected pNKs were washed twice with PBS and stained with PerCP anti-human CD45 antibody (Biolegend, San Diego, CA, USA), FITC anti-human CD3 antibody (Biolegend), BV421 anti-human CD56 antibody (Biolegend) and APC-Cy7 anti-human CD16 antibody (Biolegend) diluted in PBS with 2% FBS at 4 °C for 30 min. pNKs were then washed twice and analyzed by flow cytometry assays.
DICs isolated from decidual tissue of mice treated with DMSO or LDN57444 were collected, and the expression of CD45, CD3 and NK1.1 was analyzed by flow cytometry assays. The procedure was described above, and the antibodies used were APC-Cy7 anti-mouse CD45 antibody (Biolegend), FITC anti-mouse CD3 antibody (Biolegend) and BV421 anti-mouse NK1.1 antibody (Biolegend).
Quantitative real-time PCR
Total RNA was extracted from cells or tissues using TRIzol Reagent (Sigma-Aldrich) and subsequently reverse-transcribed into cDNA using the PrimeScript RT Master kit (TaKaRa, Shiga, Japan). Quantitative real-time PCR was performed using FastStart Universal SYBR Green Master Kits (Roche) on a ViiA7 Real-time PCR System (Applied Biosystems) according to the manufacturer’s instructions. The sequences of the primers are listed in Supplementary Table 1.
Western blot
Total proteins were extracted using lysis buffer (150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 50 mM Tris (pH 7.4), 5 mM EDTA) containing a protease inhibitor cocktail and a phosphatase inhibitor cocktail (Sigma-Aldrich). Then, protein lysates were electrophoresed via SDS‒PAGE and analyzed by western blotting with antibodies against UCHL1 (Abcam CAT# ab8189), IGFBP1 (Cell Signaling Technology, CST CAT# 31025), GAPDH (CST CAT# 2118, RRID: AB_561053), β-actin (CST CAT# 4970, RRID:AB_2223172), P-ERK1/2 (CST CAT# 4370, RRID:AB_2315112), ERK1/2 (CST CAT# 4695, RRID:AB_390779), FOXO1 (CST CAT# 2880, RRID:AB_2106495), JAK2 (CST CAT# 3230), P-STAT3 (CST CAT# 9145), and STAT3 (CST CAT# 9139).
Immunohistochemistry
Human and mouse tissue samples were fixed in 4% paraformaldehyde, embedded in paraffin after dehydration, cut at 5 µm thickness and mounted on slides. The paraffin slides were then deparaffinized and rehydrated using a graded alcohol series before H&E staining or immunohistochemistry (IHC). For IHC, the slides were boiled in 10 mM sodium citrate buffer (pH 6.0) for 10 min and then naturally cooled to room temperature. After blocking with 5% bovine serum albumin in PBS (pH 7.5), the samples were incubated overnight at 4 °C with anti-UCHL1 (Abcam CAT# ab8189), followed by incubation with an HRP-conjugated secondary antibody. Immunoreactivity was detected using a DAB kit (Gene Tech, South San Francisco, United States).
Immunofluorescence
Frozen samples of human decidual tissue, embedded in optimum cutting temperature compound (OCT, Sakura Finetek), were sectioned at 7 μm thickness for immunofluorescence. After washing with PBS, the sections were fixed with a mixture of MeOH/acetone (1:1) at − 20 °C for 5 min and blocked with 1% BSA for 1 h at 37 °C. Mouse anti-human UCHL1 (Abcam CAT# ab8189) and rabbit anti-human IGFBP1 (Abcam CAT# ab111203) were applied to slides overnight at 4 °C, followed by incubation with goat anti-mouse Alexa Fluor 488-IgG or goat anti-rabbit Alexa Fluor 555-IgG (eBioscience, California, United States) for 1 h at 37 °C in the dark. Finally, DAPI staining was performed.
Cell culture and in vitro decidualization
The immortalized human endometrial stromal cell line T-HESCs (ATCC CAT# CRL-4003TM, RRID: CVCL_C464) was a gift from Professor Haibin Wang from the Institute of Zoology, CAS (Bei**g, China)31. HESCs were cultured in DMEM/F-12 medium containing 10% FBS and 50 µg/mL penicillin/streptomycin with 1 mM sodium pyruvate, 1% insulin-transferrin-selenium (ITS), 3.1 g/L glucose, 1.5 g/L sodium bicarbonate and 500 ng/mL puromycin (all from Thermo Fisher Scientific). The culture medium was replenished every three days. In vitro decidualization of HESCs was induced as previously reported3. In brief, HESCs were treated with DMEM/F-12 containing 2% FBS, 10 µM medroxy progesterone (MPA, Selleck Chemicals, Texas, United States) and 0.5 mM 8-Br-cAMP (Selleck Chemical) for 7 days, and the medium was replenished every 2 days. DMSO (equal volume to LDN57444), LDN57444 (dissolved in DMSO at 10mM) (indicated concentrations) or C188-2 (indicated concentrations) were added during the induction of decidualization in certain experiments.
ELISA
In the in vitro decidualization assay, HESCs were treated with either DMSO or LDN57444. Then, the culture supernatant was collected at the indicated time points and centrifuged to eliminate cellular debris. The concentrations of PRL, CXCL12, IL15 and TGF-β were then analyzed according to standard protocols.
Plasmid construct and cell transfection
To construct the UCHL1 knockdown shRNA plasmid, we cloned shRNA-encoding oligonucleotides into the lentiviral pLKO.1 puro vector, a gift from Bob Weinberg (Addgene plasmid # 8453; http://n2t.net/addgene:8453; RRID: Addgene_8453) [29], to target UCHL1 mRNA. The shRNA sequences were as follows: shUCHL1-1, 5ʹ-CGGGTAGATGACAAGGTGAAT-3ʹ, shUCHL1-2, 5ʹ-GTGTGAGCTTCAGA TGGTGAA-3ʹ, shUCHL1-3′ 5ʹ-CCAGCATGAGAACTTCAGGAA-3ʹ and scramble control 5ʹ-CCTAAGGTTAAGTCGCCCTCG-3ʹ, synthesized by Sangon Biotech.
For generation of the overexpression plasmid pLVX/UCHL1, the cDNA encoding the human UCHL1 gene was amplified from the cDNA of HESCs and inserted between the XhoI and XhaI sites of pLVX-IRES-ZsGreen1 (Clontech) and pCMV-Tag2B (Stratagene). The C90S mutation of UCHL1 used PCR-based site-directed mutagenesis based on pLVX/UCHL1 with the following primers: forward, 5ʹ-AATTCCTCTGGCACAATCGGACTTATTC-3ʹ, reverse 5ʹ-CGATTGTGCCAG AGGAATTCCCAATGG-3ʹ.
To establish UCHL1 knockdown or UCHL1-overexpressing HESCs, we performed lentivirus packaging and production according to the manufacturer’s protocol (http://www.addgene.org/tools/protocols/plko/). Briefly, lentiviral plasmids harboring the desired sequences were transfected into 293T cells, together with the packing plasmids pSPAX2 and pMD2G using Lipofectamine 2000 according to the reagent protocol. The supernatants were collected 48 h after transfection, filtered through a 0.45 µm filter and used to infect HESCs in a 6-well plate supplemented with 10 µg/mL polybrene (Thermo Fisher).
Statistics
All data were shown as the mean ± SEM and were obtained from at least three independent experiments. Significant differences were analyzed by the Mann–Whitney U test, one-way ANOVA and two-way ANOVA with GraphPad Prism (version 8.0, GraphPad Software) and Statistical Package for Social Science software (version 23.0, SPSS).
Results
UCHL1 defects were related to impaired decidualization and decreased dNKs in patients with miscarriage
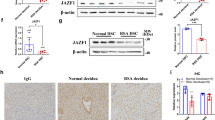
Aberrant endometrial decidualization is a major maternal cause of miscarriage [1] and frequently occurs in patients suffering pregnancy loss. Consistent with previous studies [27, 30, 31], we found that decidualization was impaired in abortion patients, as shown by abnormal morphology (Fig. 1A) and downregulation of the decidual markers IGFBP1 and PRL (Fig. 1B–D). By extension, the expression of UCHL1 was also decreased in the decidua from abortion patients (abortion decidua), as indicated by IHC staining (Fig. 1A) and immunofluorescence of UCHL1 (Fig. 1B). We then further isolated DSCs from the collected decidual tissues and examined UCHL1 expression levels. Similarly, the abortion DSCs also displayed decreased expression of UCHL1 at both the mRNA and protein levels (Fig. 1C, D).
UCHL1 defects were related to impaired decidualization and decreased dNKs in patients with miscarriage. Decidual tissues from women with normal pregnancy (normal decidua) and miscarriage (abortion decidua) were collected to determine the expression profile of UCHL1 and decidual markers. A Immunohistochemical analysis was used to verify the expression of UCHL1 in normal decidua and abortion decidua and the average optical density (AOD) analysis of UCHL-1 was analyzed. B The expression of UCHL1 and IGFBP1 was analyzed by immunofluorescence in normal decidua and abortion decidua. The mean fluorescent intensity (MFI) of UCHL1 and IGFBP1 was quantified. DSCs were then isolated from decidual tissues of women with normal pregnancy (normal DSC) and miscarriage (abortion DSC), and the expression of decidual marker genes in DSCs was detected. C The expression levels of UCHL1 and the decidual markers IGFBP1 and PRL were determined by qPCR in normal DSCs (n = 20) and abortion DSCs (n = 20). D The protein levels of UCHL1 and IGFBP1 were determined by western blotting in normal DSCs (n = 6) and abortion DSCs (n = 7). E Twenty-four hours before mRNA was collected, normal DSCs were treated with LDN57444 (5 μM). The expression of CXCL12, IL-15 and TGF-β in normal DSCs, LDN57444-treated normal DSCs and abortion DSCs was then measured by qPCR. F The culture medium (CM) of normal DSCs, LDN57444-treated normal DSCs and abortion DSCs was collected. Then, these culture media were placed into the basolateral chambers of transwell inserts, with unconditioned medium (LDN57444 added or not) as the negative control, and the pNKs were placed in the apical chambers of the transwell system. Forty-eight hours after coculture, the transwell inserts were removed, pNKs that migrated into the basolateral chamber were counted, and the statistical analysis is shown. G, H pNKs were cocultured with normal DSCs, LDN57444-treated normal DSCs and abortion DSCs, with unconditioned medium (LDN57444 added or not) as the negative control, and 48 h later, the suspended cells in the coculture system were collected for flow cytometric analysis. The proportion of CD56brightCD16− dNKs is shown in the red box (G), and the statistical analysis is shown in H. The results of the coculture assay are representative of four independent experiments and are represented by the mean ± SEM. Significant differences were determined by Mann‒Whitney U test (C, D) or one-way ANOVA (E, F, H) and are expressed as *P < 0.05, **P < 0.01, ***P < 0.001 and n.s. no significance
Additionally, we found that CXCL12, but not CXCL9, CXCL10, or CXCL11 (Supplementary Fig. 1), which are critical chemokines expressed by DSCs for the recruitment of pNKs into the fetal-maternal interface [6], was significantly downregulated in abortion DSCs compared to normal DSCs (Fig. 1E). As a result, the ability of abortion DSCs to recruit pNKs was reduced (Fig. 1F). We then measured the expression of IL15 and TGF-β, which are key cytokines for the dNKs education [32], in DSCs from different samples and found that they were also downregulated in abortion DSCs (Fig. 1E). Consequently, the percentage of CD56brightCD16− dNKs cocultured with abortion DSCs was significantly lower than that of dNKs cocultured with normal DSCs (Fig. 1G, H). Nevertheless, when pretreated with LDN57444, an inhibitor of UCHL1 [17], the expression of CXCL12, IL15 and TGF-β in normal DSCs was significantly decreased, as well as their ability to recruit and educate dNKs, which were similar to that of the abortion DSCs (Fig. 1E–H).
Collectively, these results indicated that UCHL1 was downregulated in the decidua from abortion patients, and the defect of UCHL1 was associated with the impaired decidualization and aberrant modulation of dNKs.
Functional restriction of UCHL1 disrupted HESC decidualization and consequently reduced cytokines
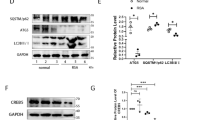
To further affirm the function of UCHL1 in human decidualization, given the difficulty of collecting human decidual tissues at different stages, we employed an immortalized human endometrial stromal cell line (T-HESC) to establish the well-known in vitro model of decidualization [33]. In this model, HESCs underwent extensive stromal to decidual transformation in response to a cocktail of cAMP plus MPA for seven days, which mimics the decidual process in vivo. The model was constructed successfully, as indicated by increased expression of the decidual markers IGFBP1 and PRL at both the mRNA level (Fig. 2A, B) and protein level (Fig. 2D, E). Excitingly, UCHL1 showed progressively increased during HESC decidualization (Fig. 2C, D), indicating a positive regulatory role of UCHL1 in decidualization.
Functional restriction of UCHL1 disrupted HESC decidualization and consequently reduced cytokines. The effect of the UCHL1 inhibitor was determined in an in vitro model of decidualization. A–C qPCR analysis showed IGFBP1, PRL and UCHL1 expression during the decidualization of HESCs. D The protein levels of UCHL1 and IGFBP1 were analyzed by western blotting. E The concentration of PRL in the culture medium of HESCs undergoing decidualization was analyzed by ELISAs. F, G HESCs were treated with DMSO or LDN57444 (5 μM) on the day that decidualization was induced. The expression of the decidual markers IGFBP1 and PRL was detected by qPCR at D1, D3, D5 and D7 of decidualization. H The concentration of PRL was determined in DMSO- or LDN57444-treated HESC culture medium by ELISAs at D1, D3, D5 and D7 of decidualization. I, J The protein levels of IGFBP1 and UCHL1 in DMSO- or LDN57444-treated HESCs were detected via western blotting at D1, D3, D5 and D7 of decidualization, with D0 as the blank control, and the statistical analysis of relative IGFBP1 expression is shown in J. K The expression of CXCL12, IL15 and TGF-β by DMSO- or LDN57444-treated HESCs at D1, D3, D5 and D7 of decidualization was detected by qPCR. L The culture media of DMSO- or LDN57444-treated HESCs at D1, D3, D5 and D7 of decidualization were collected, and the concentrations of CXCL12, IL15 and TGF-β in the media were determined by ELISAs. The results are representative of three to four independent experiments and are represented by the mean ± SEM. Significant differences were analyzed by one-way ANOVA (A–C, F) or two-way ANOVA (F–H, J–L) and are expressed as *P < 0.05, **P < 0.01, ***P < 0.001 and n.s. no significance
We then treated HESCs with the UCHL1 inhibitor LDN57444 during simulated in vitro decidualization to further verify its regulatory function in decidualization. The results showed that LDN57444 significantly reduced the mRNA levels of IGFBP1 and PRL in a dose-dependent manner (Supplementary Fig. 2A, B). Since 5 µM LDN57444 effectively inhibited decidualization of HESCs without causing substantial cell death, this concentration was used to subsequent experiments. We observed a significant decrease in the mRNA expression levels of the decidual markers IGFBP1 and PRL in the LDN57444-treated HESCs during simulated in vitro decidualization (Fig. 2F, G), as well as the other two decidual markers BMP2 and WNT 4[4] (Supplementary Fig. 2C). Moreover, western blots (Fig. 2I, J) demonstrated a decrease in IGFBP1 protein level upon treatment with LDN57444, while ELISAs displayed the downregulated expression of PRL in LDN57444-treated HESCs (Fig. 2H).
According to the results from patient samples, UCHL1 deficiency disrupted the decidual process, which further led to the decreased expression of CXCL12, IL15 and TGF-β (Fig. 1). Then, we wondered whether UCHL1 suppression also impaired chemokines and cytokines secretion in decidualized HESCs. As the data shown, mRNA levels of CXCL12, IL15 and TGF-β increased during the simulated in vitro decidualization. However, blockade of UCHL1 with LDN57444 also prevented the increase in these cell factors (Fig. 2K). Consistently, reduced concentrations of CXCL12, IL15 and TGF-β were detected in the culture medium of LDN57444-treated decidualized HESCs (Fig. 2L).
These data confirmed the role of UCHL1 in maintaining successful decidualization and expression of cell factors by decidualized HESCs.
Genetic deletion of UCHL1 induced decidualization failure along with aberrant education of dNKs
Despite being more sensitive to UCHL1 inhibition, LDN57444 also suppresses the activity of UCHL3 [26]. In addition, STAT3 signaling regulates the expression of CXCL12 [48] and TGF-β [49], cell factors secreted by DSCs to recruit and educate dNKs. In our study, we found that JAK2/STAT3 signaling was activated during decidualization and inhibited when UCHL1 was blocked by an inhibitor or shRNA. Conversely, UCHL1 overexpression enhanced the activation of JAK2/STAT3 signaling during the decidual process. Moreover, after inhibition of STAT3 using its specific inhibitor, the increased expression of IGFBP1, CXCL12, IL15 and TGF-β resulting from UCHL1 overexpression was abolished. These results suggest that the promotion of UCHL1 on decidualization and modulation of dNKs by DSCs is dependent on the activation of JAK2/STAT3 signaling.
UCHL1 is a deubiquitinating enzyme, and as its name implies, the function of UCHL1 relies on its C-terminal hydrolase to remove ubiquitin from proteins and thus inhibit protein degradation via the proteasome [35]. To verify whether the role of UCHL1 in decidualization was dependent on its deubiquitinating enzymatic activity, we generated a mutated UCHL1 (C90S) lacked deubiquitinating functionality [50] and found that the decidualization induced in vitro was significantly inhibited, so was the production of CXCL12, IL15 and TGF-β. As a result, the percentage of dNKs was also decreased when cocultured with decidualized UCHL1(C90S)-HESCs. These data suggest that the deubiquitinating ability of UCHL1 is essential for its function in endometrial decidualization. However, it remains unclear which molecule is stabilized by UCHL1 during the process of promoting decidualization. As previously reported, during decidualization, E2 elicits uterine stromal IGF1 to activate STAT3 in the epithelium, thus promoting epithelial transformation into the decidual type [51]. This research suggests that UCHL1 may regulate decidualization by stabilizing IGF1 signaling and subsequently activating activates STAT3. Another hypothesis derives from the high affinity of UCHL1 for the monomeric ubiquitin-like molecule NEDD8 [36], a monomeric ubiquitin-like molecule, as neddylation is proposed to be essential for endometrial decidualization mediated by NEDD8 [33], indicating that UCHL1 may promote decidualization through regulating neddylation. Further investigations are warranted to elucidate and validate the precise molecules or signaling pathways involved in UCHL1-mediated regulation of decidualization.
Conclusions
In conclusion, our findings demonstrated that UCHL1 deficiency was a critical cause of miscarriage, which was due to aberrant decidualization and impaired dNK modulation. The present study not only enriches the basic knowledge of the etiology of miscarriage but also provides a new intervention target to diagnose and treat miscarriage.
Availability of data and materials
The RNA-seq datasets of mouse endometrium samples generated during the current study are available in the supplementary information.
References
Quenby S, Gallos ID, Dhillon-Smith RK, Podesek M, Stephenson MD, Fisher J, et al. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet. 2021;397:1658–67.
Eschenbach DA. Treating spontaneous and induced septic abortions. Obstet Gynecol. 2015;125:1042–8.
Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012;18:1754–67.
Gellersen B, Brosens JJ. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr Rev. 2014;35:851–905.
Dunn CL, Kelly RW, Critchley HOD. Decidualization of the human endometrial stromal cell: an enigmatic transformation. Reprod Biomed Online. 2003;7:151–61.
Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol. 2013;31:387–411.
Wu X, ** L-P, Yuan M-M, Zhu Y, Wang M-Y, Li D-J. Human first-trimester trophoblast cells recruit CD56brightCD16- NK cells into decidua by way of expressing and secreting of CXCL12/stromal cell-derived factor 1. J Immunol. 2005;175:61–8.
Vacca P, Vitale C, Montaldo E, Conte R, Cantoni C, Fulcheri E, et al. CD34+ hematopoietic precursors are present in human decidua and differentiate into natural killer cells upon interaction with stromal cells. Proc Natl Acad Sci U S A. 2011;108:2402–7.
Keskin DB, Allan DSJ, Rybalov B, Andzelm MM, Stern JNH, Kopcow HD, et al. TGFbeta promotes conversion of CD16+ peripheral blood NK cells into CD16- NK cells with similarities to decidual NK cells. Proc Natl Acad Sci U S A. 2007;104:3378–83.
Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79.
Rape M. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol. 2018;19:59–70.
Fraile JM, Campos-Iglesias D, Rodríguez F, Astudillo A, Vilarrasa-Blasi R, Verdaguer-Dot N, et al. Loss of the deubiquitinase USP36 destabilizes the RNA helicase DHX33 and causes preimplantation lethality in mice. J Biol Chem. 2018;293:2183–94.
Zhang Q, Chen Q, Lu X, Zhou Z, Zhang H, Lin H-Y, et al. CUL1 promotes trophoblast cell invasion at the maternal-fetal interface. Cell Death Dis. 2013;4: e502.
Wang J, Ding J, Zhang S, Chen X, Yan S, Zhang Y, et al. Decreased USP2a Expression Inhibits Trophoblast Invasion and Associates With Recurrent Miscarriage. Front Immunol. 2021;12: 717370.
Asadpor U, Totonchi M, Sabbaghian M, Hoseinifar H, Akhound MR, Zari Moradi S, et al. Ubiquitin-specific protease (USP26) gene alterations associated with male infertility and recurrent pregnancy loss (RPL) in Iranian infertile patients. J Assist Reprod Genet. 2013;30:923–31.
Lv S, Liu M, Xu L, Zhang C. Downregulation of decidual SKP2 is associated with human recurrent miscarriage. Reprod Biol Endocrinol. 2021;19:88.
Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A, et al. Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006;126:775–88.
Liu Z, Meray RK, Grammatopoulos TN, Fredenburg RA, Cookson MR, Liu Y, et al. Membrane-associated farnesylated UCH-L1 promotes alpha-synuclein neurotoxicity and is a therapeutic target for Parkinson’s disease. Proc Natl Acad Sci U S A. 2009;106:4635–40.
Zetterberg M, Sjölander A, von Otter M, Palmér MS, Landgren S, Minthon L, et al. Ubiquitin carboxy-terminal hydrolase L1 (UCHL1) S18Y polymorphism in Alzheimer’s disease. Mol Neurodegener. 2010;5:11.
Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Kim H-J, et al. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28:117–27.
Brinkmann K, Zigrino P, Witt A, Schell M, Ackermann L, Broxtermann P, et al. Ubiquitin C-terminal hydrolase-L1 potentiates cancer chemosensitivity by stabilizing NOXA. Cell Rep. 2013;3:881–91.
Gu Y, Ding X, Huang J, Xue M, Zhang J, Wang Q, et al. The deubiquitinating enzyme UCHL1 negatively regulates the immunosuppressive capacity and survival of multipotent mesenchymal stromal cells. Cell Death Dis. 2018;9:459.
Yang D, Lu Q, Peng S, Hua J. Ubiquitin C-terminal hydrolase L1 (UCHL1), a double-edged sword in mammalian oocyte maturation and spermatogenesis. Cell Prolif. 2023;56: e13347.
Hao L, Song D, Zhuang M, Shi Y, Yu L, He Y, et al. Gene UCHL1 expresses specifically in mouse uterine decidual cells in response to estrogen. Histochem Cell Biol. 2020;154:275–86.
Telgmann R, Gellersen B. Marker genes of decidualization: activation of the decidual prolactin gene. Hum Reprod Update. 1998;4:472–9.
Zhou M, Xu H, Zhang D, Si C, Zhou X, Zhao H, et al. Decreased PIBF1/IL6/p-STAT3 during the mid-secretory phase inhibits human endometrial stromal cell proliferation and decidualization. J Adv Res. 2021;30:15–25.
Huang J, Xue M, Zhang J, Yu H, Gu Y, Du M, et al. Protective role of GPR120 in the maintenance of pregnancy by promoting decidualization via regulation of glucose metabolism. EBioMedicine. 2019;39:540–51.
Wu H-X, ** L-P, Xu B, Liang S-S, Li D-J. Decidual stromal cells recruit Th17 cells into decidua to promote proliferation and invasion of human trophoblast cells by secreting IL-17. Cell Mol Immunol. 2014;11:253–62.
Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501.
Lucas ES, Vrljicak P, Muter J, Diniz-da-Costa MM, Brighton PJ, Kong C-S, et al. Recurrent pregnancy loss is associated with a pro-senescent decidual response during the peri-implantation window. Commun Biol. 2020;3:37.
Lucas ES, Dyer NP, Murakami K, Lee YH, Chan Y-W, Grimaldi G, et al. Loss of endometrial plasticity in recurrent pregnancy loss. Stem Cells. 2016;34:346–56.
Vinketova K, Mourdjeva M, Oreshkova T. Human decidual stromal cells as a component of the implantation niche and a modulator of maternal immunity. J Pregnancy. 2016;2016:1–17.
Liao Y, Jiang Y, He H, Ni H, Tu Z, Zhang S, et al. NEDD8-mediated neddylation is required for human endometrial stromal proliferation and decidualization. Hum Reprod. 2015;30:1665–76.
Liu Y, Lashuel HA, Choi S, **ng X, Case A, Ni J, et al. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003;10:837–46.
Hurst-Kennedy J, Chin L-S, Li L. Ubiquitin C-terminal hydrolase L1 in tumorigenesis. Biochem Res Int. 2012;2012: 123706.
Wada H, Kito K, Caskey LS, Yeh ET, Kamitani T. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun. 1998;251:688–92.
Ramathal CY, Bagchi IC, Taylor RN, Bagchi MK. endometrial decidualization: of mice and men. Semin Reprod Med. 2010;28:17–26.
Fullerton PT, Monsivais D, Kommagani R, Matzuk MM. Follistatin is critical for mouse uterine receptivity and decidualization. Proc Natl Acad Sci U S A. 2017;114:E4772–81.
Ashkar AA, Black GP, Wei Q, He H, Liang L, Head JR, et al. Assessment of requirements for IL-15 and IFN regulatory factors in uterine NK Cell differentiation and function during pregnancy. J Immunol. 2003. https://doi.org/10.4049/jimmunol.171.6.2937.
Cheng HL, Schneider SL, Kane CM, Gollnick SO, Grande C, Thompson D, et al. TGF-beta 2 gene and protein expression in maternal and fetal tissues at various stages of murine development. J Reprod Immunol. 1993;25:133–48.
Lu H, Yang H-L, Zhou W-J, Lai Z-Z, Qiu X-M, Fu Q, et al. Rapamycin prevents spontaneous abortion by triggering decidual stromal cell autophagy-mediated NK cell residence. Autophagy. 2021;17:2511–27.
Farren J, Jalmbrant M, Falconieri N, Mitchell-Jones N, Bobdiwala S, Al-Memar M, et al. Posttraumatic stress, anxiety and depression following miscarriage and ectopic pregnancy: a multicenter, prospective, cohort study. Am J Obstet Gynecol. 2020;222:367.e1-367.e22.
Chen F, Sugiura Y, Myers KG, Liu Y, Lin W. Ubiquitin carboxyl-terminal hydrolase L1 is required for maintaining the structure and function of the neuromuscular junction. Proc Natl Acad Sci U S A. 2010;107:1636–41.
Rai R, Regan L. Recurrent miscarriage. Lancet. 2006;368:601–11.
Arck PC, Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring’s health. Nat Med. 2013;19:548–56.
Moffett-King A. Natural killer cells and pregnancy. Nat Rev Immunol. 2002;2:656–63.
Kitaya K, Yasuda J, Yagi I, Tada Y, Fushiki S, Honjo H. IL-15 expression at human endometrium and decidua. Biol Reprod. 2000;63:683–7.
Liu G-X, Wei J-Y, Liu M, Shi W-J, Song L-H, Li S, et al. Epigenetic upregulation of hippocampal CXCL12 contributes to context spatial memory-associated morphine conditioning. Brain Behav Immun. 2020;84:72–9.
Fu X-L, Duan W, Su C-Y, Mao F-Y, Lv Y-P, Teng Y-S, et al. Interleukin 6 induces M2 macrophage differentiation by STAT3 activation that correlates with gastric cancer progression. Cancer Immunol Immunother. 2017;66:1597–608.
Larsen CN, Price JS, Wilkinson KD. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry. 1996;35:6735–44.
Zhou C, Lv M, Wang P, Guo C, Ni Z, Bao H, et al. Sequential activation of uterine epithelial IGF1R by stromal IGF1 and embryonic IGF2 directs normal uterine preparation for embryo implantation. J Mol Cell Biol. 2021;13:646–61.
Acknowledgements
The authors thank Professor Haibin Wang (Institute of Zoology, Chinese Academy of Sciences) for his kindly gift of the human endometrial stromal cells line.
Funding
This work was supported by the National Natural Science Foundation of China (82071727, 82001550, 82070191, 82001739, 82000553 and 81873447).
Author information
Authors and Affiliations
Contributions
Yanyun Zhang, Changhong Miao and Guoqiang Zhou designed the research. Jie Zhang, Mingxing Xue and Jiefang Huang performed the experiments and wrote the paper. Shan He assisted with the collection of human samples. Lingqiao Zhu and **aonan Zhao assisted with experiments. Bei Wang and Tingwang Jiang assisted with the data analysis. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Medical Research Ethics Committee of the Affiliated Changshu Hospital of Nantong University, Changshu, China. Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, J., Xue, M., Huang, J. et al. Deficiency of UCHL1 results in insufficient decidualization accompanied by impaired dNK modulation and eventually miscarriage. J Transl Med 22, 478 (2024). https://doi.org/10.1186/s12967-024-05253-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-024-05253-0