Abstract
Background
Long noncoding RNAs (lncRNAs) have emerged as critical players in cancer progression, but their functions in colorectal cancer (CRC) metastasis have not been systematically clarified.
Methods
lncRNA expression profiles in matched normal and CRC tissue were checked using microarray analysis. The biological roles of a novel lncRNA, namely RP11-138 J23.1 (RP11), in development of CRC were checked both in vitro and in vivo. Its association with clinical progression of CRC was further analyzed.
Results
RP11 was highly expressed in CRC tissues, and its expression increased with CRC stage in patients. RP11 positively regulated the migration, invasion and epithelial mesenchymal transition (EMT) of CRC cells in vitro and enhanced liver metastasis in vivo. Post-translational upregulation of Zeb1, an EMT-related transcription factor, was essential for RP11-induced cell dissemination. Mechanistically, the RP11/hnRNPA2B1/mRNA complex accelerated the mRNA degradation of two E3 ligases, Siah1 and Fbxo45, and subsequently prevented the proteasomal degradation of Zeb1. m6A methylation was involved in the upregulation of RP11 by increasing its nuclear accumulation. Clinical analysis showed that m6A can regulate the expression of RP11, further, RP11 regulated Siah1-Fbxo45/Zeb1 was involved in the development of CRC.
Conclusions
m6A-induced lncRNA RP11 can trigger the dissemination of CRC cells via post-translational upregulation of Zeb1. Considering the high and specific levels of RP11 in CRC tissues, our present study paves the way for further investigations of RP11 as a predictive biomarker or therapeutic target for CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC), also known as large bowel cancer, is a major public health problem worldwide [1]. Epidemiological data have revealed that the 5-year survival rate of CRC patients ranges from 90% for patients with stage I disease to 10% for those with metastatic disease [2]. Although numerous studies have revealed that alterations in oncogenes and tumour suppressor genes contribute to tumorigenesis and the development of CRC [3], the precise molecular mechanisms underlying CRC pathogenesis, particularly for metastasis, remain to be fully elucidated.
Long noncoding RNAs (lncRNAs), which are more than 200 nt in length and have limited or no protein-coding capacity, play both oncogenic and tumour suppressor roles in tumorigenesis and progression [4, 5]. LncRNAs can regulate gene expression via multiple mechanisms, including chromatin remodelling, modulation of the activity of transcriptional regulators, and posttranscriptional modifications [5]. Dysregulated lncRNA expression has been reported to modulate the progression of various types of cancers, such as bladder, prostate, lung, breast, gastric and colorectal cancers [6, 7]. Increasing evidence suggests that lncRNAs can trigger metastatic progression, increase chromosomal instability, and promote CRC tumorigenesis [8,Full size image
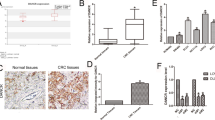
Among the eight candidate genes, targets of the CUST_8502_PI428631609 (lncRNA RP11, RP11-138 J23.1) and CUST_9335_PI428631609 (lncRNA AC123023.1) probes have been shown to be lncRNAs. Microarray analysis suggested that the elevation in lncRNA RP11 (RP11) in CRC tissues versus adjacent normal tissues was greater than that of lncRNA AC123023.1 (Table S3). qRT-PCR confirmed that the abundance of RP11 was significantly greater than that of lncRNA AC123023.1 in 5 CRC tissues (Additional file 1: Figure S1 C). RP11 located at Chromosome 5: 104,079,911-104,105,403 with the transcript length 574 nt (ENSG00000251026, Additional file 1: Figure S1 B). It was poly A-tailed due to the enrichment in bound fractions was 11-fold greater than that in unbound fractions by use of polydT-beads pull down and qRT-PCR.
To confirm the role of RP11 in the progression of CRC, we compared RP11 levels in CRC tissues and paired adjacent non-cancerous mucosa from 32 individual patients (Table S1). RP11 was successfully amplified in all tumour and normal specimens analysed. According to the qRT-PCR analysis, RP11 expression was significantly increased in 30 out of 32 (93.8%) tumour samples compared with the adjacent normal mucosa tissues (Fig. 1 d). In this cohort, the average expression level of RP11 in the tumour tissues was 48-fold greater than that in the adjacent normal mucosa tissues. However, there was no significant difference in RP11 expression between different ages, sexes or stages (Table S1), which might be due to the small sample size.
We further assessed RP11 expression in a TCGA pan-cancer dataset obtained from the GEPIA online database (http://gepia.cancer-pku.cn). TCGA data confirmed that the expression of RP11 in colon and rectal carcinoma (COAD, READ) was significantly (p < 0.05) greater than that in the adjacent normal tissues (Fig. 1 e). In addition, the expression of RP11 in COAD and READ was relatively high among all measured cancers (Additional file 1: Figure S1 D and E). RP11 expression was verified in multiple colon cancer cell lines, namely, SW620, LoVo, HCT-116, Caco2, HT29, HCT-15, HCT-8, SW480, DLD1, and RKO, and in human colon mucosal epithelial NCM460 cells. The results indicated that the RP11 levels in all of the measured CRC cell lines, except RKO, were greater than that in NCM460 cells (Fig. 1 f). SW620 cells, which were primarily derived from lymph node metastases in CRC patients, had the highest level of RP11 among all analysed cell lines (Fig. 1 f). Collectively, these data show that lncRNA RP11 is increased in CRC cells and tissues.
RP11 triggers the dissemination of CRC cells both in vitro and in vivo
The potential biological roles of RP11 in CRC progression were investigated. We overexpressed RP11 in HCT-15, HCT-8, DLD1, SW480 and RKO cells (RP11 low expression cells, Additional file 1: Figure S2 A). CCK-8 analysis showed RP11 overexpression had no significant effect on the proliferation of these cells (Fig. 2 a). Consistently, RP11 silencing in SW620 or HCT-116 cells (RP11 high expression cells, Additional file 1: Figure S2 B) also had no significant effect on cell proliferation (Additional file 1: Figure S2 C). The colony formation analysis showed that RP11 overexpression had no significant effect on colony formation of HCT-15 or HCT-8 cells (Additional file 1: Figure S2 D). Flow cytometry showed that RP11 overexpression had no significant effect on HCT-15 or HCT-8 cell cycle progression (Additional file 1: Figure S2 E). In addition, RP11 overexpression had no significant effect on stress-induced apoptosis, doxorubicin sensitivity, rhodamine123 efflux or ROS generation in HCT-15 (Additional file 1: Figure S2 F~I) or HCT-8 (data not shown) cells.
RP11 triggers the dissemination of CRC cells both in vitro and in vivo. a CRC cells were transfected with the vector control or pcDNA/RP11 for 48 h, and proliferation was measured with a CCK-8 kit. b The wound healing of HCT-15 RP11 stable overexpression and control cells was recorded (left) and quantitatively analysed (right). c The in vitro invasion of HCT-15 RP11 stable overexpression and control cells was recorded (left) and quantitatively analysed (right). d The expression of EMT-related markers of HCT-15 or HCT-8 RP11 stable overexpression and control cells was verified by western blot analysis. e After transfection with si-NC or si-RP11 for 48 h, the expression of EMT markers in SW620 cells was verified by western blot analysis. f Tumour growth curves of HCT-15 RP11 stable overexpression and control cells in xenograft models at the indicated time intervals. g Weights of tumours derived from HCT-15 RP11 stable overexpression or control cells in xenograft models at the end of the experiment. h IHC analysis of Ki67-, vimentin- or fibronectin-stained paraffin-embedded sections obtained from xenografts. (I).HCT-15 RP11 stable overexpression and control cells were injected into nude mice via the tail vein. Representative images and H&E staining of metastatic liver tumours are shown. j The number of metastatic sites of tumours derived from HCT-15 RP11 stable overexpression or control cells was quantitatively analysed. Data are presented as the mean ± SD from three independent experiments. Bar = 200 μm. *p < 0.05, **p < 0.01 compared with control
The effects of RP11 on the in vitro migration and invasion of CRC cells were evaluated. A wound healing analysis revealed that RP11 overexpression triggered the migration of both HCT-15 (Fig. 2 b) and HCT-8 (Additional file 1: Figure S2 J) cells. Transwell analysis confirmed that RP11 can increase the in vitro invasion of HCT-15 cells (Fig. 2 c). RP11 silencing inhibited the in vitro migration (Additional file 1: Figure S2 K) and invasion (Additional file 1: Figure S2 L) of SW620 cells. CRC cells overexpressing RP11 assumed their spindle-like fibroblast appearance and lost their cobblestone-like epithelial morphology (Additional file 1: Figure S2 M), suggesting that RP11 may regulate EMT and cancer metastasis. This was confirmed by western blot analysis, which showed a decrease in the expression of epithelial cell marker E-Cadherin (E-Cad) and an increase in the expression of mesenchymal cell markers fibronectin (FN) and Vimentin (Vim) in HCT-15 and HCT-8 cells transfected with RP11 (Fig. 2 d). RP11 silencing impaired EMT progression in SW620 (Fig. 2 e) and HCT-116 (Additional file 1: Figure S2 N) cells. Collectively, our data suggested that RP11 can induce the migration, invasion and EMT of CRC cells.
To evaluate the in vivo effects of RP11 on tumour development, we examined the expression levels of EMT-related markers in RP11-overexpressing HCT-15 tumour xenografts in nude mice. At the end of the experiment, the tumour sizes, volumes and weights in the RP11 group were comparable to those in the control group (Fig. 2 f, g). This was confirmed by IHC analysis of the expression of Ki67, a nuclear antigen expressed in proliferating cells, and the Ki67 level was comparable between the RP11 and control groups (Fig. 2 h). The IHC data showed that RP11 increased the levels of Vim and FN in HCT-15 tumour xenografts (Fig. 2 h).
To further determine the impacts of RP11 on in vivo metastasis, equal numbers of HCT-15 RP11 stable overexpression and control cells (1 × l06 in 100 μl) were injected into BALB/c nude mice via the tail vein, and distant liver metastasis was analysed. Eight weeks after injection, the experiment was terminated, and the liver was analysed for the presence of metastatic tumours. As shown in Fig. 2 i & j, the numbers and sizes of the liver tumours derived from RP11-overexpressing HCT-15 cells were significantly greater than those derived from the control cells. Collectively, our data showed that RP11 can enhance the in vitro and in vivo dissemination of CRC cells and induce EMT.
Upregulation of Zeb1 mediates the RP11-induced dissemination of CRC cells
LncRNA can activate the transcription of closely located genes in cis by promoting chromatin loo** from transcriptional enhancers [25, 26]. We therefore investigated the effects of RP11 on its nearby transcripts, including NUDT12, C5orf30, PPIP5K2, GIN1, RP11-6 N13.1, and CTD-2374C24 (Additional file 1: Figure S1 B). The expression levels of the detected genes showed no significant difference between the HCT-15 RP11 stable and control cells (Additional file 1: Figure S3 A). In SW620 cells, RP11 knockdown also had no effect on the expression of its nearby transcripts (Additional file 1: Figure S3 B). Thus, the biological functions of RP11 may not be related to the cis regulatory function.
EMT-TFs such as Snail, Slug, Twist and Zeb1 can regulate the progression of EMT by targeting E-Cad expression [27]. To investigate the mechanisms responsible for the RP11-induced dissemination of CRC cells, we analysed the effects of RP11 on the expression of EMT-TFs in CRC cells. The results showed that RP11 overexpression increased the expression of Zeb1 in both HCT-15 and HCT-8 cells, while si-RP11 downregulated the expression of Zeb1 in SW620 and HCT-116 cells (Fig. 3 a and Additional file 1: Figure S3 C). RP11 overexpression or knockdown had no effect on the expression of Snail, Slug or Twist (Fig. 3 a and Additional file 1: Figure S3 C). The subcellular fraction showed that RP11 overexpression increased the nuclear accumulation of Zeb1 in HCT-15 cells (Fig. 3 b). Consistently, RP11 increased Zeb1 expression in HCT-15 tumour xenografts (Fig. 3 c). Intriguingly, neither RP11 overexpression in HCT-15 (Fig. 3 d) nor knockdown in SW620 (Additional file 1: Figure S3 D) cells had significant effect on the mRNA levels of tested EMT-TFs. Consistently, RP11 overexpression had no effect on the mRNA expression of Zeb1 in Caco2, HT-29, SW480, DLD1, or RKO cells (Additional file 1: Figure S3 E).
Upregulation of Zeb1 mediates the RP11-induced dissemination of CRC cells. a. The expression levels of EMT-TFs in HCT-15 or HCT-8 RP11 stable overexpression and control cells were verified by western blot analysis. After transfection with si-NC or si-RP11 for 48 h, the expression levels of EMT-TFs in SW620 cells were verified by western blot analysis. b Zeb1 expression in subcellular fractions of HCT-15 RP11 stable overexpression and control cells was verified by western blot analysis. c IHC analysis of Zeb1-stained paraffin-embedded sections obtained from xenografts. d The mRNA expression levels of EMT-TFs in HCT-15 RP11 stable overexpression and control cells were verified by qRT-PCR. e After transfection with si-NC or si-Zeb1 for 48 h, the wound healing of HCT-15 RP11 stable overexpression and control cells was quantitatively analysed. f After transfection with si-NC or si-Zeb1 for 48 h, the EMT markers of HCT-15 RP11 stable overexpression and control cells were detected by western blot analysis. g After treatment with 100 μg/ml CHX for the indicated times, Zeb1 expression in HCT-15 RP11 stable overexpression and control cells was detected by western blot analysis (left) and quantitatively analysed (right). h & i Zeb1 in HCT-15 or HCT-8 RP11 stable overexpression and control cells was immunoprecipitated for the detection of ubiquitylation. Data are presented as the mean ± SD from three independent experiments. Bar = 200 μm. **p < 0.01 compared with control
Although Zeb1 has been well demonstrated to induce the EMT of cancer cells, including CRC cells, by inhibiting E-Cad [17, 28], the role of Zeb1 in the RP11-induced dissemination of CRC cells was unknown and thus investigated. A wound healing analysis showed that Zeb1 knockdown attenuated RP11-induced cell migration (Fig. 3 e, Additional file 1: Figure S3 F). Western blot analysis confirmed that Zeb1 knockdown attenuated RP11-induced upregulation of FN and downregulation of E-Cad (Fig. 3 f).
The results indicated that RP11 may increase Zeb1 expression via post-translational regulation. This was confirmed by data showing that the half-life of the Zeb1 protein in HCT-15 (Fig. 3 g) and HCT-8 (Additional file 1: Figure S3 G) RP11 stable overexpression cells was significantly greater than that in their corresponding control cells. Because ubiquitylation of Zeb1 is critical for its stabilization [29], we hypothesized that RP11 modified the ubiquitylation level of Zeb1. Immunoprecipitation results showed that RP11 can significantly decrease the ubiquitylation of Zeb1 in both HCT-15 (Fig. 3 h) and HCT-8 (Fig. 3 i) cells. Collectively, our present data suggested that the post-translational upregulation of Zeb1 is involved in the RP11-induced dissemination of CRC cells.
Downregulation of Siah1 and Fbxo45 mediates RP11-induced upregulation of Zeb1
Because lncRNAs can directly intact with proteins and thereby regulate protein stability [25, 30], the binding of Zeb1 to RP11 was investigated by RIP-PCR. The data showed that immunoprecipitation (IP) of Zeb1 had no significant effect on RP11 recruitment in either HCT-15 or HCT-8 cells (Additional file 1: Figure S4 A). In addition, Zeb1 overexpression had no effect on the RP11 expression in either HCT-15 or HCT-8 cells (Additional file 1: Figure S4 B). Consistently, the RP11 pull-down/MS analysis did not show binding between RP11 and Zeb1 in either the HCT-15 control or RP11 stable overexpression cells (Table S4). This suggested that the RP11-induced upregulation of Zeb1 is not due to a direct interaction. GSK-3β, β-catenin, p65, MAPK/ERK, p38-MAPK, PI3K/Akt, and STAT3 have been reported to regulate Zeb1 expression and EMT [31]. However, no significant variation was observed in the total and phosphorylated levels of these signalling molecules between HCT-15 RP11 stable overexpression and control cells (Additional file 1: Figure S4 C).
To systematically investigate the specific factors involved in the RP11-induced stabilization of Zeb1 in CRC cells, we examined the mRNA expression levels of 7 reported factors in the ubiquitin–proteasome system, which can post-translationally regulate the stability of Zeb1 (summarized in Table S5). The results indicated that RP11 overexpression significantly (p < 0.05) decreased the expression levels of Siah1 and Fbxo45 but had no significant effect on other factors in either HCT-15 (Fig. 4 a) or HCT-8 (Fig. 4 b) cells. This was confirmed by a western blot analysis showing that RP11 overexpression downregulated the expression of Siah1 and Fbxo45 in both HCT-15 and HCT-8 cells (Fig. 4 c). Consistently, RP11 decreased the expression of Siah1 and Fbxo45 in HCT-15 tumour xenografts (Fig. 4 d).
Downregulation of Siah1 and Fbxo45 mediates the RP11-induced upregulation of Zeb1. a & b The mRNA expression levels of 7 reported target proteins related to Zeb1 stability in HCT-15 a or HCT-8 (b) RP11 stable overexpression and control cells were determined by qRT-PCR. c The protein expression of Siah1 and Fbxo45 in HCT-15 or HCT-8 RP11 stable overexpression and control cells was determined by western blot analysis. d IHC analysis of Siah1- or Fbxo45-stained paraffin-embedded sections obtained from HCT-15 RP11 stable overexpression and control xenografts. e HCT-15 cells were transfected with vector control, pcDNA/RP11, pcDNA/Siah1, or Fbxo45 alone or together for 48 h, protein expression was verified by western blot analysis. f RIP-PCR was performed to analyse the relative enrichment of RP11 by use of an antibody against Siah1 or Fbxo45 in HCT-15 cells. Data are presented as the means ± SD from three independent experiments. Bar = 200 μm. **p < 0.01 compared with control
To verify the roles of Siah1 and Fbox45 in the expression of Zeb1, we overexpressed Siah1 and Fbxo45 in HCT-15 cells (Fig. 4 e). The results showed that overexpression of Siah1 and Fbxo45 attenuated the RP11-induced upregulation of Zeb1 in HCT-15 cells (Fig. 4 e). However, RIP-PCR showed that RP11 had no significant effect on the recruitment of Siah1 or Fbxo45 protein in HCT-15 cells (Fig. 4 f). Consistently, the RP11 pull-down/MS analysis did not show binding between RP11 and Siah1 or Fbxo45 in HCT-15 cells (Table S4). This result suggested that RP11 downregulates the mRNA levels of Siah1 and Fbxo45 but does not bind to the Siah1 or Fbxo45 protein.
RP11 regulates Siah1 and Fbxo45 expression by forming the RP11-hnRNPA2B1-mRNA complex
To investigate the potential mechanisms of the RP11-regulated mRNA expression of Siah1 and Fbxo45, we performed RNA pull-down assays followed by MS with biotinylated RP11 and antisense RP11 as a negative control. Among the identified proteins summarized in Table S4, hnRNPA2B1 was identified as a protein that directly interacted with RP11 (Fig. 5 a) and has been reported to shorten mRNA half-lives [32]. RIP analysis verified the interaction between hnRNPA2B1 and RP11 in HCT-15 cells (Fig. 5 b&c).
RP11 regulates Siah1 and Fbxo45 expression by forming the RP11-hnRNPA2B1-mRNA complex. a RNA pull-down analysis and MS identified hnRNPA2B1 as the specific protein interacting with RP11 in both HCT-15 and HCT-15 RP11 stable overexpression cells. The red arrow shows the position of hnRNPA2B1. b The secondary structure of RP11 was predicted (http://rna.tbi.univie.ac.at/). The red colour indicates strong confidence for the prediction of each base. c RNA pull-down detection of the interaction between hnRNPA2B1 and RP11, Siah1, or Fbxo45 in HCT-15 cells. d hnRNPA2B1 expression in the cytoplasmic and nuclear fractions of HCT-15 RP11 stable overexpression and control cells were analysed by western blot. e HCT-15 cells were transfected with pcDNA (vector) or pcDNA/hnRNPA2B1 for 24 h, and the expression of Siah1 and Fbxo45 was verified by western blot analysis. f &g The computational prediction of the interaction between RP11 and the Siah1 (f) or Fbxo45 (g) mRNA based on IntaRNA 2.0 (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) [53]. h After in vitro transcription to generate biotin-labelled RP-11 and RP-11 AS, RIP-PCR was performed to analyse the relative enrichment of Siah1 or Fbxo45 mRNA on RP11 in HCT-15 cells. i & j After treatment with Act-D for the indicated times, the mRNA levels of Siah1 (i) or Fbxo45 (j) in HCT15 RP11 stable overexpression and control cells were measured by qRT-PCR. k Binding between hnRNPA2B1 and Siah1 mRNA or between hnRNPA2B1 and Fbxo45 mRNA in HCT-15 RP11 stable overexpression and control cells was analysed by RIP-PCR. Data are presented as the mean ± SD from three independent experiments. **p < 0.01 compared with control
hnRNPA2B1 is an RNA binding protein (RBP) and localizes in both the cytoplasm and nucleus. Our data showed that RP11 overexpression increased the cellular localization of hnRNPA2B1 in the cytoplasm in both HCT-15 (Fig. 5 d) and HCT-8 (Additional file 1: Figure S5 A) cells. RIP-PCR showed that hnRNPA2B1 could recruit both Siah1 and Fbxo45 mRNA in HCT-15 cells (Fig. 5 c). Furthermore, hnRNPA2B1 overexpression decreased the mRNA (Additional file 1: Figure S5 B) and protein (Fig. 5 e) expression of Siah1 and Fbxo45 in HCT-15 cells.
Computational analysis revealed that RP11 could directly bind to the CDS of Siah1 (Fig. 5 f) and the 3’UTR of Fbxo45 (Fig. 5 g). In vitro transcription and RIP-PCR confirmed that RP11 could directly bind to the mRNA of Siah1 and Fbxo45 in HCT-15 cells (Fig. 5 h). RP11 overexpression significantly downregulated the mRNA stability of Siah1 (Fig. 5 i) and Fbxo45 (Fig. 5 j) in HCT-15 cells.
We further investigated whether binding between hnRNPA2B1 and the mRNA of Siah1 and Fbxo45 was RP11 dependent. RIP-PCR showed that the binding between hnRNPA2B1 and the mRNA of Siah1 and Fbxo45 in the HCT-15 RP11 stable overexpression cells was significantly greater than that in the control cells (Fig. 5 k). Consistently, RP11 knockdown decreased the binding between hnRNPA2B1 and the mRNA of Siah1 and between hnRNPA2B1 and the mRNA of Fbxo45 in HCT-15 cells (Additional file 1: Figure S5 C). These data suggested that RP11 regulates Siah1 and Fbxo45 expression by forming the RP11-hnRNPA2B1-mRNA complex.
m6A modification is involved in the upregulation of RP11 in CRC cells
The epigenetic mechanisms responsible for the upregulation of RP11 in CRC cells were investigated. First, treatment with 5-aza-dC (a DNA methyltransferase inhibitor) had no significant effect on RP11expression in either HCT-15 or HCT-8 cells (Additional file 1: Figure S6 A), suggesting that DNA methylation might not be involved in RP11 expression in CRC cells. The role of histone acetylation in RP11 expression was investigated by treating HCT-15 cells with specific inhibitors of HDAC1, 3, 4, 6 and 8 or broad-spectrum HDAC inhibitors such as SAHA and NaB. The data showed that these HDAC inhibitors had no significant effect on RP11 expression in HCT-15 cells (Additional file 1: Figure S6 B). This was confirmed by data showing that overexpression of HDAC6 and HDAC8 had no effect on RP11 expression in HCT-15 cells (Additional file 1: Figure S6 C).
The N6-methyladenosine (m6A) modification modulates all stages of the RNA life cycle, such as RNA processing, nuclear export and translation [33, 34], and thereby regulates the expression and functions of RNAs, including lncRNAs. m6A RNA-immunoprecipitation (RIP) qPCR showed 9.3- and 5.0-fold enrichment in m6A antibody levels of RP11 in HCT-15 and HCT-8 cells, respectively (Fig. 6 a), while the level of enrichment (2.3-fold) in NCM460 cells was significantly less than that in CRC cells (Fig. 6 a). We found that overexpression of Mettl3 (Additional file 1: Figure S6 D), the key m6A methyltransferase (“writer”) in mammalian cells [35, 36], increased RP11 expression in both HCT-15 and HCT-8 cells (Fig. 6 b). Consistently, overexpression of ALKBH5 (Additional file 1: Figure S6 E), the demethylase of m6A, decreased RP11 expression (Fig. 6 c). These data indicated that m6A positively regulates RP11 expression in CRC cells.
The m6A modification is involved in the upregulation of RP11 in CRC cells. a m6A RIP-qPCR analysis of RP11 in HCT-15, HCT-8 and NCM460 cells. b After transfection with vector control or ppB/Mettl3 for 24 h, RP11 expression was measured by qRT-PCR. c After transfection with vector control or pcDNA/Alkbh5 for 24 h, RP11 expression was measured by qRT-PCR. d After transfection with vector control or ppB/Mettl3 for 24 h, HCT-15 cells were further treated with Act-D for the indicated times, and RP11 expression was measured by qRT-PCR. e After transfection with vector control or ppB/Mettl3 for 24 h, the cytoplasmic, nuclear, and chromatin fractions of HCT-15 cells were separated for RNA extraction and qRT-PCR. f After transfection with vector control or ppB/Mettl3 for 24 h, binding between RP11 and hnRNPA2B1 in HCT-15 and HCT-8 cells was analysed by RIP-PCR using an antibody against hnRNPA2B1. Data are presented as the mean ± SD from three independent experiments. *p < 0.05, **p < 0.01 compared with control
We then evaluated the possible mechanisms involved in the m6A-regulated expression of RP11 in CRC cells. By treating cells with Act-D to terminate transcription, our data revealed that Mettl3 overexpression had no significant effect on the half-life of RP11 in HCT-15 cells (Fig. 6 d). The results of subcellular fractionation analysis showed that Mettl3 overexpression could markedly increase the localization of RP11 to chromatin (Fig. 6 e), which might be because Mettl3 can increase the stability of nascent RP11. However, Mettl3 overexpression had no effect on the mRNA expression of Siah1 or Fbxo45 in HCT-15 cells (Additional file 1: Figure S6 F). Furthermore, Mettl3 overexpression increased binding between RP11 and hnRNPA2B1 in both HCT-15 and HCT-8 cells (Fig. 6 f), which might be due to Mettl3 increasing RP11 expression and hnRNPA2B1 is a m6A reader for the RNA processing events [37]. These data suggested that the m6A modification can increase RP11 expression in CRC cells by increasing RP11 nuclear accumulation.
The m6A/RP11/Zeb1 axis and in vivo progression of CRC
At this point, we asked whether there was a link between m6A methylation-regulated RP11, its downstream molecules Siah1, Fbxo45, and Zeb1, and clinical CRC development. Zeb1 expression in CRC tissues was significantly (p < 0.01) greater than that in normal tissues, according to Hong Colorectal (Fig. 7 a) and Skrzypczak Colorectal 2 data (Fig. 7 b) from the Oncomine database. Significantly increased Zeb1 was observed in patients with N2 stage CRC compared to patients with N0 stage CRC (Fig. 7 c). Consistently, decreased expression of Fbxo45 was observed in patients with N2 stage CRC compared to patients with N0 stage CRC (Fig. 7 d). In addition, Mettl3 expression in patients with N2 stage CRC was significantly greater than that in patients with N1 stage CRC (Additional file 1: Figure S7 A). However, there was no significant difference for Siah1 between patients with N0, N1 or N2 stage CRC (Additional file 1: Figure S7 B). This finding suggested an increasing trend for METTL3 and Zeb1 expression and a decreasing trend for Fbxo45 expression during the malignant transformation of CRC. We further verified the co-expression relationship for RP11-regulated CRC progression. We found that RP11 expression was significantly negatively correlated with ALKBH5 expression in CRC patients (Additional file 1: Figure S7 C). This confirmed that the m6A can regulate the expression of RP11, further, RP11 regulated Siah1-Fbxo45/Zeb1 was involved in the development of CRC.
The m6A/RP11/Zeb1 axis and in vivo progression of CRC. a & b The relative mRNA expression of Zeb1 in two Oncomine datasets: Hong Colorectal (a), and Skrzypczak Colorectal 2 (b). c & d The relative mRNA expression of Zeb1 (c) and Fbxo45 (d) in patients with stage N0, N1, and N2 CRC based on data available from TCGA database. e DFS of CRC patients with high (n = 135) and low (n = 134) levels of RP11 was plotted according to the Kaplan-Meier method. f DFS of CRC patients with high (n = 135) and low (n = 135) levels of RP11/Siah1 was plotted according to the Kaplan-Meier method. g DFS of CRC patients with high (n = 135) and low (n = 135) levels of RP11/Fbxo45 was plotted according to the Kaplan-Meier method. (H) DFS of CRC patients with high (n = 135) and low (n = 134) levels of Zeb1 was plotted according to the Kaplan-Meier method. (I) DFS of CRC patients with high (n = 135) and low (n = 135) levels of Zeb1/Siah1 was plotted according to the Kaplan-Meier method. j DFS of CRC patients with high (n = 135) and low (n = 135) levels of Zeb1/Fbxo45 was plotted according to the Kaplan-Meier method. *p < 0.05, **p < 0.01 compared with control
Using the online Kaplan-Meier plotter bioinformatics tool, we found that colon cancer patients with increased RP11 expression showed reduced disease-free survival (DFS, Fig. 7 e) and overall survival (OS, Additional file 1: Figure S7 D), with no significant difference (p > 0.05). When RP11 expression was normalized to that of Siah1 (the relative levels of RP11 to that of Siah1) or Fbxo45, there was a trend towards significance for the reduced DFS of colon patients with higher RP11/Siah1 (Fig. 7 f) or RP11/Fbxo45 levels (Fig. 7 g) compared with patients with lower values. Similarly, there was a trend towards significance for the reduced OS of colon cancer patients with higher RP11/Siah1 (Additional file 1: Figure S7 E) or RP11/Fbxo45 levels (Additional file 1: Figure S7 F) compared with those with lower values.
We found that colon cancer patients with increased Zeb1 expression showed reduced DFS (Fig. 7 h) with significant difference (p < 0.05). When Zeb1 expression was normalized to that of Fbxo45 (Fig. 7 j), but not Siah1 (Fig. 7 i), the DFS of colon cancer patients with higher Zeb1/Siah1 levels (Fig. 7 j) was statistically significantly reduced compared to patients with lower values. Similarly, colon cancer patients with increased Zeb1 expression showed significantly (p < 0.05) reduced OS compared with patients with the low levels (Additional file 1: Figure S7 G). Normalization to Fbxo45 (Additional file 1: Figure S7 I), but not Siah1 (Additional file 1: Figure S7 J), further significantly reduced the OS of colon cancer patients. These results suggest that the m6A/RP11/Zeb1 axis triggers the in vivo progression of CRC.