Abstract
Background
As the rupture of cerebral aneurysm may lead to fatal results, early detection of unruptured aneurysms may save lives. At present, the contrast-unenhanced time-of-flight magnetic resonance angiography is one of the most commonly used methods for screening aneurysms. The computer-assisted detection system for cerebral aneurysms can help clinicians improve the accuracy of aneurysm diagnosis. As fully convolutional network could classify the image pixel-wise, its three-dimensional implementation is highly suitable for the classification of the vascular structure. However, because the volume of blood vessels in the image is relatively small, 3D convolutional neural network does not work well for blood vessels.
Results
The presented study developed a computer-assisted detection system for cerebral aneurysms in the contrast-unenhanced time-of-flight magnetic resonance angiography image. The system first extracts the volume of interest with a fully automatic vessel segmentation algorithm, then uses 3D-UNet-based fully convolutional network to detect the aneurysm areas. A total of 131 magnetic resonance angiography image data are used in this study, among which 76 are training sets, 20 are internal test sets and 35 are external test sets. The presented system obtained 94.4% sensitivity in the fivefold cross-validation of the internal test sets and obtained 82.9% sensitivity with 0.86 false positive/case in the detection of the external test sets.
Conclusions
The proposed computer-assisted detection system can automatically detect the suspected aneurysm areas in contrast-unenhanced time-of-flight magnetic resonance angiography images. It can be used for aneurysm screening in the daily physical examination.
Similar content being viewed by others
Background
Among people without comorbidity, with an average age of 50 years, the prevalence of unruptured intracranial aneurysms is about 3.2% in a population without comorbidity [1]. Though it has a strong latency, some aneurysms may show no symptoms for years or even decades, the rupture of one aneurysm may lead to serious neurological sequelae and may be fatal. Under such circumstances, the prediction of when an aneurysm will rupture becomes very important. Hence, an automated detection system for cerebral aneurysms may help clinicians in the earlier and more accurate diagnosis of aneurysms. TOF-MRA as a non-invasive imaging technique shows promising diagnostic accuracy compared with DSA, which is the gold standard diagnostic method for aneurysm [2]. Therefore, TOF-MRA is currently one of the most commonly used methods for screening aneurysms, of which 3.0 T is the most popular [3].
Deep neural networks have been used to detect cerebral aneurysms since 2017 [4,5,6,7,8,9]. Up until now, several methods have been proposed in this field [4,5,6,7,8,9,10,11,12,13,3).
Development of the CAD system
In our system, we designed two main steps: first, automatic segmentation of the artery vascular voxels; second, aneurysm detection based on deep neural networks. And there were two pipelines, one for the training of the neural network model, as the preparation of the system, the other for the actual detection of the aneurysms in the real data and show the results to users. So the clinicians only need to input DICOM image sets and then the system would show where the aneurysms were with a high likelihood (Fig. 4).
Step one: segmentation
The input was DICOM datasets, which is in the form of a volume. In view of the many three-dimensional features of cerebral arteries, our method processed the image data as volume from start to finish. In the segmentation step, the input image data were pre-processed using N3 bias field correction and histogram normalization. Then the vessel region was enhanced using a sigmoid filter, the total transform is given by the formula below, with α = 400 and β = 600:
Seed points on skull were selected automatically based on the bounding-box method. This method used a cube to wrap the skull from the outside and shrank it. When the faces of the cube came into contact with the skull, the contact points were used as the seed points. The auto-threshold region was allowed to grow from the seed points and smooth the results, then the skull region voxels were obtained. The lower threshold of the region-growing method was 30% of the maximum intensity, and upper threshold was the maximum intensity. These voxels were cut off from the pre-processed data and the skull was removed. Since the high signal area in TOF-MRA data was mainly skull and vessels, artery blood vessel accounted for a large part of the left high signal objects. We binarized the data based on the intensity region of the vessel, set all voxels greater than the background density value to 1. Then we performed connected domain statistics, arranged connected domains according to the number of voxels it contains, and selected seed points from the 5 connected domains that are ranked first. In most instances, the selected seed area was the main branch of artery vessels. Since the intensity of the vessels in TOF-MRA data followed the Gaussian distribution [22], the upper and lower thresholds of the region-growing method were decided automatically as μ + σ and μ − σ (μ stands for the estimated mean and σ stands for the standard deviation). With the automatic region growth, vessel voxels were segmented from the TOF-MRA data, reconstructed the surface of the vessel using the marching cubes method, then the surface mesh model of artery vessels was obtained. The mesh model could be used in the hemodynamic analysis since it was the inner surface of vessels. The target of the CAD system was the detection of aneurysms; to do the detection, the vessel voxels were dilated using radius 10 sphere kernel, and the vessel area and its neighbor area were the input of step two. The auto-segmented vessel area output of step one covered 100% of the labeled aneurysms in our dataset (Fig. 5).
Step two: detection
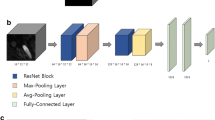
In this step, we used a deep neural network to detect the aneurysms in the image. Since the blood vessel was a continuous structure in three dimensions, and usually had a long radial length, traditional 2D CNNs could not be applied on the important 3D structure features, which were widely used in clinical diagnosis. Fully convolutional networks (FCNs), such as U-Net, made full use of every pixel in the image and brought semantic segmentation to a practical level. In this paper, we chose an improved 3D-UNet [23] as our detection method. The structure of this network was below. Inspired by U-Net, this network could process 3D input blocks of 128*128*128 voxels, and also comprised a context aggregation pathway as U-Net. Besides, they employed deep supervision to the network by injecting gradient signals.
To train our model, we augmented the 96 image sets to 768, using flip** (by transverse section), histogram normalization, discrete Gaussian noise filter (variance: 4.0, max kernel width: 32 pixels, sequentially. The image sets were then cropped and resampled to 128*128*128. The annotations were dilated based on the center of the labeled area, all annotations were dilated to a sphere with the same radius. The way to choose the radius was above 3 voxels under 128*128*128, which was above 3% of the length of axis. After the dilation, the annotations had two labeled objects, the vessel area as background, and aneurysms as foreground. Then 76 of the 96 image sets were selected as training datasets and put into the network for training. The initial factors were: batch size = 1, initial learning rate = 5e−4, optimization function was Adam, the weights were initialized using the default initializer (glorot_uniform) of Keras. After about 200 epochs the learning process got an early stop; it costs 10 h in our environment.
After training, we obtained the deep neural network model and used it to predict the new TOF-MRA image; the image was processed by step one, got the vessel area, then the model would predict each voxel in the vessel area. The model would give the likelihood of each voxel to be normal vessel or aneurysm. These possibilities were binarized at a threshold of 0.5, a value greater than 0.5 was converted to 1 and a value less than 0.5 was converted to 0. Then each voxel of the vessel area was classified into 2 labels: vessel area and aneurysm area. The voxels of one label were all connected to be one component, so we took the center of the aneurysm area and draw a sphere from this point, with the radius the same as used to dilate the annotations. The area inside the sphere had a high probability to be an aneurysm, and clinicians could check the area more carefully to make the diagnosis (Fig. 6).
Availability of data and materials
Not applicable.
Abbreviations
- TOF-MRA:
-
Time-of-flight magnetic resonance angiography
- DSA:
-
Digital subtraction angiography
- CNN:
-
Convolutional neural networks
- FCN:
-
Fully convolutional network
- CAD:
-
Computer-assisted detection
- CPU:
-
Computer-assisted detection
- RAM:
-
Random access memory
- GPU:
-
Graphic processing unit
- MCA:
-
Middle cerebral artery
- PCA:
-
Posterior cerebral artery
- ICA:
-
Internal carotid artery
- ACA:
-
Anterior cerebral artery
- BA:
-
Basilar artery
- VA:
-
Vertebral artery
- FP:
-
False-positive cases
- DICOM:
-
Digital Imaging and Communications in Medicine (DICOM) is the standard for the communication and management of medical imaging information and related data
References
Vlak MH, Algra A, Brandenburg R, Rinkel GJ. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. Lancet Neurol. 2011;10(7):626–36.
Sailer AM, Wagemans BA, Nelemans PJ, de Graaf R, van Zwam WH. Diagnosing intracranial aneurysms with MR angiography: systematic review and meta-analysis. Stroke. 2014;45(1):119–26.
Kaufmann TJ, Huston JI, Cloft HJ, Mandrekar J, Gray L, Bernstein MA, Atkinson JL, Kallmes DF. A prospective trial of 3T and 1.5T time-of-flight and contrast-enhanced MR angiography in the follow-up of coiled intracranial aneurysms. Am J Neuroradiol. 2010;31(5):912–8.
El Hamdaoui H, Maaroufi M, Alami B, Chaoui N, Boujraf S. Computer-aided diagnosis systems for detecting intracranial aneurysms using 3D angiographic data sets. In: 2017 international conference on advanced technologies for signal and image processing (ATSIP). IEEE; 2017, p. 1–5.
Moccia S, De Momi E, El Hadji S, Mattos LS. Blood vessel segmentation algorithms—review of methods, datasets and evaluation metrics. Comput Methods Programs Biomed. 2018;158:71–91.
Nakao T, Hanaoka S, Nomura Y, Sato I, Nemoto M, Miki S, Maeda E, Yoshikawa T, Hayashi N, Abe O. Deep neural network-based computer-assisted detection of cerebral aneurysms in MR angiography. J Magn Reson Imaging. 2018;47(4):948–53.
Nemoto M, Hayashi N, Hanaoka S, Nomura Y, Miki S, Yoshikawa T. Feasibility study of a generalized framework for develo** computer-aided detection systems—a new paradigm. J Digit Imaging. 2017;30(5):629–39.
Ueda D, Yamamoto A, Nishimori M, Shimono T, Doishita S, Shimazaki A, Katayama Y, Fukumoto S, Choppin A, Shimahara Y. Deep learning for MR angiography: automated detection of cerebral aneurysms. Radiology. 2018;290(1):187–94.
Yu L, Cheng J-Z, Dou Q, Yang X, Chen H, Qin J, Heng P-A. Automatic 3D cardiovascular MR segmentation with densely-connected volumetric convnets. In: International conference on medical image computing and computer-assisted intervention. Springer; 2017, p. 287–95.
Hanaoka S, Nomura Y, Takenaga T, Murata M, Nakao T, Miki S, Yoshikawa T, Hayashi N, Abe O, Shimizu A. HoTPiG: a novel graph-based 3-D image feature set and its applications to computer-assisted detection of cerebral aneurysms and lung nodules. Int J Comput Assist Radiol Surg. 2019;14:2095–107.
Hamdaoui HE, Maaroufi M, Alami B, Chaoui NE, Boujraf S. Computer-aided diagnosis systems for detecting intracranial aneurysms using 3D angiographic data sets: Review. In: International conference on advanced technologies for signal and image processing; 2017, p. 1–5.
Hentschke CM, Beuing O, Paukisch H, Scherlach C, Skalej M, Tönnies KD. A system to detect cerebral aneurysms in multimodality angiographic data sets. Med Phys. 2014;41(9):1904.
Malik KM, Anjum SM, Soltanian-Zadeh H, Malik H, Malik GM. A framework for intracranial saccular aneurysm detection and quantification using morphological analysis of cerebral angiograms. IEEE Access. 2018;6:7970–86.
**ao R, Ding H, Zhai F, Zhou W, Wang G. Cerebrovascular segmentation of TOF-MRA based on seed point detection and multiple-feature fusion. Comput Med Imag Graph. 2018;69:1–8.
Duan H, Huang Y, Liu L, Dai H, Chen L, Zhou L. Automatic detection on intracranial aneurysm from digital subtraction angiography with cascade convolutional neural networks. Biomed Eng Online. 2019;18(1):1–18.
Sabour S, Li Z-Y. Reproducibility of image-based computational models of intracranial aneurysm; methodological issue. Biomed Eng Online. 2016;15(1):109.
Wong KKL, Wang D, Ko JKL, Mazumdar J, Le T-T, Ghista D. Computational medical imaging and hemodynamics framework for functional analysis and assessment of cardiovascular structures. Biomed Eng Online. 2017;16(1):35.
Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2014;39:640–51.
Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. Ar**v 2015, abs/1505.04597.
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Medical image computing and computer-assisted intervention; 2016, p. 424–32.
Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Transac Pattern Anal Mach Intell. 2018;40(4):834–48.
Wen L, Wang X, Wu Z, Zhou M, ** JS. A novel statistical cerebrovascular segmentation algorithm with particle swarm optimization. Neurocomputing. 2015;148:569–77.
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH. Brain tumor segmentation and radiomics survival prediction: contribution to the brats 2017 challenge. In: International MICCAI Brainlesion Workshop. Springer; 2017, p. 287–97.
Acknowledgements
Not applicable.
Funding
This work was supported by National Key Research and Development Plan (2018YFC0116904), National Natural Science Foundation of China (61672236), Jiangsu Key Technology Research Development Program (BE2017663), Suzhou Industry Technological Innovation Projects (SYG201707), Suzhou Science and Technology Development Project (SZS201818), Lishui Key Technology Research Development Program (2019ZDYF09, 2019ZDYF17).
Author information
Authors and Affiliations
Contributions
GC suggested the CAD system for cerebral aneurysm. GC and XW implemented it and analyzed the images. HL and LY acquired and annotated the MR angiography images. DY and GD reviewed the results of the imaging diagnosis. YL was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The ethics board of Huashan Hospital comprehensively reviewed and approved the protocol of this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chen, G., Wei, X., Lei, H. et al. Automated computer-assisted detection system for cerebral aneurysms in time-of-flight magnetic resonance angiography using fully convolutional network. BioMed Eng OnLine 19, 38 (2020). https://doi.org/10.1186/s12938-020-00770-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12938-020-00770-7