Abstract
Background
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease with neuronal cell inclusions composed of neurofilaments and other abnormal aggregative proteins as pathological hallmarks. Approximately 90% of patients have sporadic cases (sALS), and at least 4 genes, i.e. C9orf72, SOD1, FUS and TARDBP, have been identified as the main causative genes, while many others have been proposed as potential risk genes. However, these mutations could explain only ~ 10% of sALS cases. The neurofilament polypeptides encoded by NEFH, NEFM, and NEFL are promising protein biomarkers for ALS and other degenerative diseases. However, whether the genetic variants of these genes were associated with ALS remain ambiguous.
Methods
Here, we used PCR-Sanger to sequence the exons of these three genes in a cohort of 371 sALS patients and 711 healthy controls (Phase I) and validated the risk variant in another 300 sALS patients and 1076 controls (Phase II).
Results
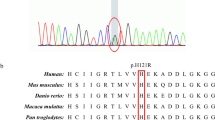
A total of 92 variants were identified, including 36 rare heterozygous variants in NEFH, 27 in NEFM, and 16 in NEFL, and only rs568759161 (p.Ser787Arg) in NEFH reached nominal statistical power (P = 0.02 at Phase I, P = 0.009 at Phase II) in the case–control comparison. Together, the Phase I and II studies showed the significantly higher frequency of the variant in cases (9/1342, 0.67%) than in controls (2/3574, 0.07%) (OR 12.06; 95% CI 2.60–55.88; P = 0.0003). No variants passed multiple testing in the discovery cohort, but rs568759161 was associated with ALS in a replication cohort.
Conclusions
Our results confirmed that NEFH Ser787Arg is a novel sALS risk variant in Chinese subjects, but NEFM and NEFL were not associated with sALS. These data may have implications for genetic counselling and for understanding the pathogenesis of sALS.
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease characterized by loss of motor neurons in the brain and spinal cord, resulting in muscle atrophy, swallowing disorders, and pyramidal tract signs. A known pathological hallmark of ALS is neuronal cell inclusions composed of neurofilaments and other abnormal aggregative proteins [1,2,3]. Epidemiological surveys show an incidence of 0.6–3.8 per 100,000 persons per year and a prevalence of 4.1–8.4 per 100,000 persons worldwide [4]. It has been reported that the yearly incidence is 0.8 (2010–2015) per 100,000 persons in China [5]. Approximately 10% of cases were familial, and 90% were sporadic cases. To date, the genetics of ALS are not fully understood. In 1993, SOD1 was discovered as the first ALS-causing gene. Since then, many other genes have been reported to be causative for (i.e. C9orf72, SOD1, FUS, TARDBP, etc.) or associated with the disease [6, 7]. Genetic studies have found that mutations in these genes were mainly identified in familial cases and could explain only approximately 10% of sporadic cases (sALS) [6]. With next-generation sequencing, novel genes and loci have been increasingly discovered [40], and motor neuron disease in mice [32]. NEFM is linked to Parkinson's disease [41]. However, NEFL and NEFM were not associated with sALS in our study. Although we had shown variants in these genes in sALS, we did not find significant differences in clinical characteristics (sex, AAO, onset site) between cases carrying and not carrying the variants. The differences further confirmed the genetic heterogeneity in sALS among different ethnicities and highlight the association of NEFH, but not NEFL or NEFM, with ALS.
This study found that the p.Ser787Arg variant in NEFH was associated with sALS in Chinese subjects. Notably, rs568759161 is only found in only the East Asian population according to ExAC (MAF 0.14%) and gnomAD (MAF 0.24%), and their MAFs were slightly higher than those of our control group (0.07% in Phase I and 0.05% in Phase II) (Table 3). We assume that the difference in allele frequency between the two databases and our study might be due to the population differences. Moreover, we found that some variants reported by other studies were not associated with ALS. For example, A380T in NEFH was identified only in cases previously [29], but our study suggested it was identified in both case and control groups. So, we believed study of rare variants need large samples of controls. In our study, we recruited relatively large controls (n = 711 in Phase I and n = 1076 in Phase II) to decrease the chance of false positive or false negative.
The phosphorylation of NF subunits has been considered a critical process regulating the formation and function of NFs [10]. The variant p.Ser787Arg is located in the phosphorylated region in a conserved sequence. Proper phosphorylation/dephosphorylation of NEFH may be considered a protective mechanism under conditions of cellular stress [16, 42], indicating that the modification of NEFH plays a significant role in maintaining the normal function of neurons. We hypothesized that the NEFH-S787R variant changes the phosphorylation of the protein. However, because of the unavailability of an antibody against the site, we did not test the hypothesis in this study. In the future, we need to synthesize antibodies against the phosphorylated NEFH-Ser787 site to further explore the changes in phosphorylation levels.
Recently, next-generation sequencing technology have identified many genes, including NEFH [43], as causative for or associated with ALS. In NEFH, 20 variants have been reported in ALS cases (Fig. 1) [25, 36,37,38, 44, 45], but none was conclusively related to the disease. The p.Ser787Arg was only reported by Chen et al. [45], but its association with ALS was not confirmed. Our study provided the spectrum of NEFH variants and confirmed the association of p. Ser787Arg with Chinese sALS.
Conclusion
In this study, we analysed the mutational spectrum of NEFH, NEFM, and NEFL genes in an sALS Chinese cohort and identified the variant (rs568759161) locating in the phosphorylated site of the KSP domain of NEFH as a risk variant associated with sALS in Chinese. Functional studies will be necessary to assess its role in ALS pathogenesis.
Availability of data and materials
All genetic polymorphisms identified in this study is available at Table 2, Additional file 2: Table S2 and Additional file 5: Table S5. The primer sequences are shown in Additional file 1: Table S1. The original sequencing and clinical datasets generated during the current study are not publicly available due to maintaining patient confidentiality but are available from the corresponding author (hh-p@163.com) on reasonable request.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- NFs:
-
Neurofilaments
- FTD:
-
Frontotemporal dementia
- AAO:
-
Age at onset
- PCR:
-
Polymerase chain reaction
- ExAC:
-
Exome aggregation consortium
- MAFs:
-
Minor allele frequencies
- CADD:
-
Combined annotation-dependent depletion
- SKAT-O:
-
Sequence kernel association test
- LoF:
-
Loss-of-function
References
Leung CL, He CZ, Kaufmann P, Chin SS, Naini A, Liem RK, et al. A pathogenic peripherin gene mutation in a patient with amyotrophic lateral sclerosis. Brain Pathol. 2004;14:290–6.
McAlary L, Plotkin SS, Yerbury JJ, Cashman NR. Prion-like propagation of protein misfolding and aggregation in amyotrophic lateral sclerosis. Front Mol Neurosci. 2019;12:262.
Lin H, Schlaepfer WW. Role of neurofilament aggregation in motor neuron disease. Ann Neurol. 2006;60:399–406.
Longinetti E, Fang F. Epidemiology of amyotrophic lateral sclerosis: an update of recent literature. Curr Opin Neurol. 2019;32:771–6.
Zhou S, Zhou Y, Qian S, Chang W, Wang L, Fan D. Amyotrophic lateral sclerosis in Bei**g: epidemiologic features and prognosis from 2010 to 2015. Brain Behav. 2018;8:e01131.
Taylor JP, Brown RH Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206.
Hardiman O, Al-Chalabi A, Chio A, Corr EM, Logroscino G, Robberecht W, et al. Amyotrophic lateral sclerosis. Nat Rev Dis Primers. 2017;3:17085.
Pampalakis G, Mitropoulos K, **romerisiou G, Dardiotis E, Deretzi G, Anagnostouli M, Katsila T, Rentzos M, Patrinos GP. New molecular diagnostic trends and biomarkers for amyotrophic lateral sclerosis. Hum Mutat. 2019;40(4):361–73.
Gentil BJ, Tibshirani M, Durham HD. Neurofilament dynamics and involvement in neurological disorders. Cell Tissue Res. 2015;360:609–20.
Al-Chalabi A, Miller CC. Neurofilaments and neurological disease. BioEssays. 2003;25:346–55.
Kobayakawa Y, Sakumi K, Kajitani K, Kadoya T, Horie H, Kira J, et al. Galectin-1 deficiency improves axonal swelling of motor neurones in SOD1(G93A) transgenic mice. Neuropathol Appl Neurobiol. 2015;41:227–44.
Hirano A, Donnenfeld H, Sasaki S, Nakano I. Fine structural observations of neurofilamentous changes in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 1984;43:461–70.
Hirano A, Nakano I, Kurland LT, Mulder DW, Holley PW, Saccomanno G. Fine structural study of neurofibrillary changes in a family with amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 1984;43:471–80.
Sasaki S, Maruyama S, Yamane K, Sakuma H, Takeishi M. Swellings of proximal axons in a case of motor neuron disease. Ann Neurol. 1989;25:520–2.
Elder GA, Friedrich VL Jr, Pereira D, Tu PH, Zhang B, et al. Mice with disrupted midsized and heavy neurofilament genes lack axonal neurofilaments but have unaltered numbers of axonal microtubules. J Neurosci Res. 1999;57:23–32.
Liu Q, **e F, Siedlak SL, Nunomura A, Honda K, Moreira PI, et al. Neurofilament proteins in neurodegenerative diseases. Cell Mol Life Sci. 2004;61:3057–75.
Ge WW, Leystra-Lantz C, Wen W, Strong MJ. Selective loss of trans-acting instability determinants of neurofilament mRNA in amyotrophic lateral sclerosis spinal cord. J Biol Chem. 2003;278:26558–63.
Bergeron C, Beric-Maskarel K, Muntasser S, Weyer L, Somerville MJ, Percy ME. Neurofilament light and polyadenylated mRNA levels are decreased in amyotrophic lateral sclerosis motor neurons. J Neuropathol Exp Neurol. 1994;53:221–30.
Poesen K, Van Damme P. Diagnostic and prognostic performance of neurofilaments in ALS. Front Neurol. 2018;9:1167.
Khalil M, Teunissen CE, Otto M, Piehl F, Sormani MP, Gattringer T, et al. Neurofilaments as biomarkers in neurological disorders. Nat Rev Neurol. 2018;14:577–89.
Oeckl P, Jardel C, Salachas F, Lamari F, Andersen PM, Bowser R, et al. Multicenter validation of CSF neurofilaments as diagnostic biomarkers for ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2016;17:404–13.
Figlewicz DA, Krizus A, Martinoli MG, Meininger V, Dib M, Rouleau GA, et al. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum Mol Genet. 1994;3:1757–61.
Skvortsova V, Shadrina M, Slominsky P, Levitsky G, Kondratieva E, Zherebtsova A, et al. Analysis of heavy neurofilament subunit gene polymorphism in Russian patients with sporadic motor neuron disease (MND). Eur J Hum Genet. 2004;12:241–4.
Al-Chalabi A, Andersen PM, Nilsson P, Chioza B, Andersson JL, Russ C, et al. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum Mol Genet. 1999;8:157–64.
Tripolszki K, Gampawar P, Schmidt H, Nagy ZF, Nagy D, Klivényi P, et al. Comprehensive genetic analysis of a Hungarian amyotrophic lateral sclerosis cohort. Front Genet. 2019;10:732.
Tomkins J, Usher P, Slade JY, Ince PG, Curtis A, Bushby K, et al. Novel insertion in the KSP region of the neurofilament heavy gene in amyotrophic lateral sclerosis (ALS). NeuroReport. 1998;9:3967–70.
Vechio JD, Bruijn LI, Xu Z, Brown RH Jr, Cleveland DW. Sequence variants in human neurofilament proteins: absence of linkage to familial amyotrophic lateral sclerosis. Ann Neurol. 1996;40:603–10.
Rooke K, Figlewicz DA, Han FY, Rouleau GA. Analysis of the KSP repeat of the neurofilament heavy subunit in familiar amyotrophic lateral sclerosis. Neurology. 1996;46:789–90.
Garcia ML, Singleton AB, Hernandez D, Ward CM, Evey C, Sapp PA, et al. Mutations in neurofilament genes are not a significant primary cause of non-SOD1-mediated amyotrophic lateral sclerosis. Neurobiol Dis. 2006;21:102–9.
Julien JP. Amyotrophic lateral sclerosis. Unfolding the toxicity of the misfolded. Cell. 2001;104:581–91.
Wu YY, Kuo HC. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J Biomed Sci. 2020;27:49.
Bomont P, Cavalier L, Blondeau F, Ben Hamida C, Belal S, Tazir M, et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet. 2000;26:370–4.
Brooks BR, Miller RG, Swash M, Munsat TL. World federation of neurology research group on motor neuron diseases: El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:293–9.
Dorst J, Chen L, Rosenbohm A, Dreyhaupt J, Hübers A, Schuster J, et al. Prognostic factors in ALS: a comparison between Germany and China. J Neurol. 2019;266:1516–25.
Yuan A, Rao MV, Veeranna Nixon RA. Neurofilaments and neurofilament proteins in health and disease. Cold Spring Harb Perspect Biol. 2017;9:a018309.
Nakamura R, Sone J, Atsuta N, Tohnai G, Watanabe H, Yokoi D, et al. Next-generation sequencing of 28 ALS-related genes in a Japanese ALS cohort. Neurobiol Aging. 2016;39(219):e1-8.
Nishiyama A, Niihori T, Warita H, Izumi R, Akiyama T, Kato M, et al. Comprehensive targeted next-generation sequencing in Japanese familial amyotrophic lateral sclerosis. Neurobiol Aging. 2017;53:194.e1-194.e8.
Morgan S, Shatunov A, Sproviero W, Jones AR, Shoai M, Hughes D, et al. A comprehensive analysis of rare genetic variation in amyotrophic lateral sclerosis in the UK. Brain. 2017;140:1611–8.
Maciel R, Correa R, Bosso Taniguchi J, Prufer Araujo I, Saporta MA. Human tridimensional neuronal cultures for phenotypic drug screening in inherited peripheral neuropathies. Clin Pharmacol Ther. 2020;107:1231–9.
Agrawal PB, Joshi M, Marinakis NS, Schmitz-Abe K, Ciarlini PD, Sargent JC, et al. Expanding the phenotype associated with the NEFL mutation: neuromuscular disease in a family with overlap** myopathic and neurogenic findings. JAMA Neurol. 2014;71:1413–20.
Lavedan C, Buchholtz S, Nussbaum RL, Albin RL, Polymeropoulos MH. A mutation in the human neurofilament M gene in Parkinson’s disease that suggests a role for the cytoskeleton in neuronal degeneration. Neurosci Lett. 2002;322:57–61.
Wataya T, Nunomura A, Smith MA, Siedlak SL, Harris PL, Shimohama S, et al. High molecular weight neurofilament proteins are physiological substrates of adduction by the lipid peroxidation product hydroxynonenal. J Biol Chem. 2002;277:4644–8.
Pecoraro V, Mandrioli J, Carone C, Chiò A, Traynor BJ, Trenti T. The NGS technology for the identification of genes associated with the ALS. A systematic review. Eur J Clin Invest. 2020;50(5):e13228.
Liu ZJ, Lin HX, Wei Q, Zhang QJ, Chen CX, Tao QQ, Liu GL, Ni W, Gitler AD, Li HF, Wu ZY. Genetic spectrum and variability in Chinese patients with amyotrophic lateral sclerosis. Aging Dis. 2019;10(6):1199–206.
Chen W, **e Y, Zheng M, Lin J, Huang P, Pei Z, Yao X. Clinical and genetic features of patients with amyotrophic lateral sclerosis in southern China. Eur J Neurol. 2020;27(6):1017–22.
Acknowledgements
We thank all the participants of this study. We’re also grateful for Dr. **uli Feng and Dr. Shu **e for their technical assistance, including PRC and Sanger sequencing.
Funding
This study was supported by funds from the Innovation of Science and Technology, Fujian Province (Grant Number 2017Y9058), to Prof. Huapin Huang; the Fujian Provincial Health Technology Project (Grant Number 2019-ZQN-38), to Dr. Wanhui Lin; the Fujian Sanming Science and Technology Plan Project (Grant Number 2019-S-3) and Fujian Provincial Science and Technology Project (Grant Number 2020J011271), to Dr. Feng Lin; and the Ministry of Science and Technology (Grant Number 2016YFC1306000), Special Fund from Key Laboratory of Neurodegenerative Diseases, Ministry of Education of China (PXM2019_026283_000002), and National Natural Science Foundation (Grant Number 81771212) to Prof. Chaodong Wang.
Author information
Authors and Affiliations
Contributions
FL and HH initiated the project and wrote the manuscript. FL, WL, and CZ designed the experiments. FL, CZ, JL, and XL performed the experiments. FL, ZW, and CW analysed the data. FL, XL, JZ and JL constructed the figures. FL, XL, JZ and XL created the tables. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All research participants or their legal representatives signed informed consent forms for participation in clinical and genetic research. The medical ethics committee approved the protocol and provided informed consent (2021KY016) of the whole study at Fujian Medical University Union Hospital, Fuzhou, China. In this study, all methods were performed in accordance with the relevant guidelines and regulations in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
None of the authors declared conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Primers for amplification of exons.
Additional file 2
. Rare non-synonymous coding variants in NEFM and NEFL.
Additional file 3
. Burden in all rare variants.
Additional file 4
. Clinical features of NEFH (S787R) carriers.
Additional file 5
. Low frequency and common variants identified in NEFH, NEFM and NEFL genes.
Additional file 6
. Workflow of the study design.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lin, F., Lin, W., Zhu, C. et al. Sequencing of neurofilament genes identified NEFH Ser787Arg as a novel risk variant of sporadic amyotrophic lateral sclerosis in Chinese subjects. BMC Med Genomics 14, 222 (2021). https://doi.org/10.1186/s12920-021-01073-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-021-01073-z