Abstract
Objective
Ubiquitin-specific peptidase 10 (USP10), a typical de-ubiquitinase, has been found to play a double-edged role in human cancers. Previously, we reported that the expression of USP10 was negatively correlated with the depth of gastric wall invasion, lymph node metastasis, and prognosis in gastric cancer (GC) patients. However, it remains unclear whether USP10 can regulate the metastasis of GC cells through its de-ubiquitination function.
Methods
In this study, proteome, ubiquitinome, and transcriptome analyses were conducted to comprehensively identify novel de-ubiquitination targets for USP10 in GC cells. Subsequently, a series of validation experiments, including in vitro cell culture studies, in vivo metastatic tumor models, and clinical sample analyses, were performed to elucidate the regulatory mechanism of USP10 and its de-ubiquitination targets in GC metastasis.
Results
After overexpression of USP10 in GC cells, 146 proteins, 489 ubiquitin sites, and 61 mRNAs exhibited differential expression. By integrating the results of multi-omics, we ultimately screened 9 potential substrates of USP10, including TNFRSF10B, SLC2A3, CD44, CSTF2, RPS27, TPD52, GPS1, RNF185, and MED16. Among them, TNFRSF10B was further verified as a direct de-ubiquitination target for USP10 by Co-IP and protein stabilization assays. The dysregulation of USP10 or TNFRSF10B affected the migration and invasion of GC cells in vitro and in vivo models. Molecular mechanism studies showed that USP10 inhibited the epithelial-mesenchymal transition (EMT) process by increasing the stability of TNFRSF10B protein, thereby regulating the migration and invasion of GC cells. Finally, the retrospective clinical sample studies demonstrated that the downregulation of TNFRSF10B expression was associated with poor survival among 4 of 7 GC cohorts, and the expression of TNFRSF10B protein was significantly negatively correlated with the incidence of distant metastasis, diffuse type, and poorly cohesive carcinoma.
Conclusions
Our study established a high-throughput strategy for screening de-ubiquitination targets for USP10 and further confirmed that inhibiting the ubiquitination of TNFRSF10B might be a promising therapeutic strategy for GC metastasis.
Similar content being viewed by others
Introduction
Ubiquitin-specific protease 10 (USP10) is a typical de-ubiquitinase that cleaves ubiquitin from ubiquitin-conjugated protein substrates to enhance protein stability [Full size image
To further characterize the de-ubiquitination substrates of USP10, we employed an affinity-based ubiquitinated peptide enrichment approach to systematically quantify the change of ubiquitinome in AGS cells with USP10 overexpression. As the tryptic peptides with TMT/iTRAQ Labeling were fractionated into fractions by high pH reverse-phase HPLC, we carried out affinity enrichment of ubiquitin modified peptides using pan antibody-based PTM enrichment. The enriched peptides were then subjected to LC-MS/MS analysis for ubiquitinome quantification. We identified 3581 ubiquitination sites on 1554 proteins, of which 3148 sites on 1434 proteins were quantifiable (Fig. 1C). With a criterion of ≥ 1.5-fold change between the study group and the control group, we finally obtained 136 significant downregulated ubiquitin sites on 123 proteins (Fig. 1C), which might directly be regulated by USP10. We used UALCAN website (https://ualcan.path.uab.edu/analysis.html) to conveniently calculate and define GC progression related genes according to gene expression and clinical samples data of TCGA (STAD). Based on their relationship with GC progression related genes, proteins deubiquitinated by USP10 can be classified into three categories (Table S3). Among them, 36 deubiquitinated sites on 29 deubiquitinated proteins could be associated with promotion metastasis, and 18 deubiquitinated sites on 17 deubiquitinated proteins could be associated with inhibiting metastasis (Fig. 1D, Table S3). To investigate whether there was any particular amino acid preference adjacent to the ubiquitinated sites, we analyzed the model of sequences constituted with amino acids near the ubiquitin sites in all protein sequences using Soft MoMo. The results showed that aspartic acid (D) and leucine (L) were notably close to the ubiquitin sites, whereas lysine (K) and arginine (R) were less preferred near the ubiquitin sites (Fig. 1E), and the motif was xxxxxxxxLx_K_xxxxxxxxxx (Motif Score = 8.69) or xxxxxxxLxx_K_xxxxxxxxxx (Motif Score = 7.37) (Fig. S1A).
To determine the direct substrates of USP10, we compared the upregulated proteins and downregulated ubiquitinated sites. Ideally, the direct substrates of USP10 were identified from two independent experiments (i.e., proteome and ubiquitinome). We considered that a protein identified from two datasets could be a potential substrate of USP10. Using this criterion, we identified 9 potential substrates of USP10, including TNFRSF10B, SLC2A3, CD44, CSTF2, RPS27, TPD52, GPS1, RNF185, and MED16 (Fig. 1F). The ubiquitinated expression levels of these 9 proteins were downregulated, while the total protein expression levels were upregulated in response to USP10 overexpression (Fig. 1G and H). Signal pathway enrichment analysis demonstrated that USP10 upregulating proteins were enriched in EMT-related GO functions and pathways, such as cell migration, epithelial cell-cell adhesion, extracellular matrix, pathways in cancer, etc. (Fig. 1I, Fig. S1B-D). Moreover, 61 differentially expressed genes were identified by transcriptome sequencing (Fig. 1J). However, none of these genes and their signal pathways overlapped with the 9 potential substrates of USP10 (Fig. 1K, Fig. S2), further indicating that these proteins were upregulated by USP10 at the level of translation or post-translational modifications. The motif analysis showed that the modified sequence of CD44 and TPD52 belonged to xxxxxxxLxx_K_xxxxxxxxxx, and the modified sequence of RPS27 and MED16 belonged to xxxxxxxxLx_K_xxxxxxxxxx.
USP10 regulates the migration and invasion of GC cells
Three siRNAs targeted USP10 mRNA were designed and transfected to AGS or MKN45 cells. Among them, the si-USP10-2 knockdown effect was the most stable (Fig. S3). Then, the result showed that down-regulation of USP10 notably enhanced the ability of migration compared with the negative control group (si-NC) (Fig. 2A). Meanwhile, the invasive cells were observably increased in the GC cells after USP10 knockdown (Fig. 2B). USP10 protein levels were markedly reduced when si-USP10 was transfected into AGS or MKN45 cells (Fig. 2C). Furthermore, to exclude the off-target effect of siRNA, three si-USP10s (40 nM) were transfected respectively. The results of wound healing showed that all these si-USP10s increased the migration ability of AGS cells (Fig. S4A). Surprisingly, when the concentration of si-USP10s increased to 60 nM, the effect of si-USP10s on cell migration was reversed (Fig. S4B).
USP10 regulates the migration and invasion of GC cells. A Wound-healing assay (left) and quantitative analysis (right) in human GC cell lines (AGS or MKN45) transfected with si-USP10, original magnification, ×40. B Transwell invasion assay (left) and quantitative analysis (right) in human GC cell lines transfected with si-USP10, original magnification, ×40. C The protein level of USP10 was measured when si-USP10 was transfected into AGS cells or MKN45 cells. D Wound-healing assay (left) and quantitative analysis (right) in human GC cell lines transfected with GV657-USP10, original magnification, ×40. E Transwell invasion assay (left) and quantitative analysis (right) in human GC cell lines transfected with GV657-USP10, original magnification, ×40. F The protein level of USP10 was measured when GV657-USP10 was transfected into AGS cells or MKN45 cells. n = 6 per group, *p < 0.05, **p < 0.01
To further verify that USP10 inhibited the migration and invasion of GC cells, we constructed overexpressed plasmid GV657-USP10. Figure 2D showed that up-regulation of USP10 significantly reduced the ability of migration compared to the negative control group. Meanwhile, the invasive cells were decreased in the group of GV657-USP10 transfected after 24 h and 48 h (Fig. 2E). USP10 protein levels were markedly increased when the GV657-USP10 was transfected into AGS or MKN45 cells (Fig. 2F).
USP10 regulates TNFRSF10B stabilization via de-ubiquitination in GC cells
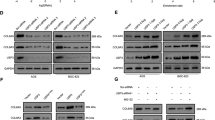
Herein, we chose TNFRSF10B and MED16 as the representatives for further verification. After transfection with si-USP10, the protein expression levels of TNFRSF10B were reduced in AGS and MKN45 cells, while the protein expression levels of MED16 were only reduced in MKN45 cells (Fig. 3A-C). In addition, the mRNA expression level of TNFRSF10B and MED16 did not change significantly after si-USP10 transfected into MKN45 cells (Fig. 3D). The Co-IP assays showed that USP10 could bind to TNFRSF10B in 293T cells transfected with GV657-USP10 (Fig. 3E), further indicating that USP10 can regulate the de-ubiquitination of TNFRSF10B. After treatment with the proteasome inhibitor MG132, TNFRSF10B protein expression was significantly increased in the GV657-USP10 group (Fig. 3F). And the cycloheximide (CHX, a protein synthesis inhibitor) assays showed that the degradation rate of TNFRSF10B protein at different time points (0, 3, 6, and 9 h) slowed down significantly in the GV657-USP10 group (Fig. 3G and H), further confirming that USP10 directly upregulated TNFRSF10B protein expression via de-ubiquitination.
USP10 regulated TNFRSF10B stabilization via de-ubiquitination in GC cells. A, B and C The protein expression level of USP10, TNFRSF10B, and MED16 were measured by WB after si-USP10 transfected into AGS cells or MKN45 cells. D The mRNA expression level of USP10, TNFRSF10B, and MED16 were measured by qRT-PCR after si-USP10 transfected into MKN45 cells. E Co-IP assay: after 293T cells were transfected with Flag-GV657-USP10 plasmid (Flag tag), the cell lysates were co-immunoprecipitated by anti-Flag (USP10) antibody with IgG as negative control. The immunoprecipitated proteins of USP10 and TNFRSF10B were detected by WB, that is, the membrane was incubated with TNFRSF10B antibody. Then, the membrane was incubated with anti-Flag (USP10) antibody. F After 20 µM MG132 treated 8 h, the protein expression level of USP10 and TNFRSF10B were measured by WB in 293T cells transfected with GV657-USP10. G, H After 10 µM cycloheximide (CHX) treated at different time points, the protein expression level of USP10 and TNFRSF10B were measured in 293T cells transfected with GV657-USP10, WB analysis (left) and densitometry analysis (right). n = 6 per group, *p < 0.05, **p < 0.01
Downregulated TNFRSF10B enhances the migration ability of GC cells in vitro
GO enrichment analysis based on proteomics analysis result showed that USP10 upregulating TNFRSF10B involvement in the cell migration signaling pathway of AGS cells (Table S4). Therefore, we further assessed the migration ability of AGS or MKN45 cells which were transfected with si-TNFRSF10B-1, si-TNFRSF10B-2, or si-TNFRSF10B-3 by the wound-healing experiments. After si-TNFRSF10B-1, si-TNFRSF10B-2, or si-TNFRSF10B-3 transfected into AGS or MKN45 cells, the migration ability of GC cells was notably elevated (Fig. 4A and B). Western blot assays demonstrated that TNFRSF10B protein levels were markedly reduced when si-TNFRSF10B-1, si-TNFRSF10B-2, or si-TNFRSF10B-3 was transfected into AGS or MKN45 cells (Fig. 4C).
Downregulated TNFRSF10B enhanced the migration ability of GC cells in vitro. A, B Wound-healing assay (left) and quantitative analysis (right) in human GC cell lines (AGS or MKN45) transfected with si-TNFRSF10B-1, si-TNFRSF10B-2, or si-TNFRSF10B-3, original magnification, ×40. C The protein level of TNFRSF10B was measured when si-TNFRSF10B-1, si-TNFRSF10B-2, or si-TNFRSF10B-3 was transfected into AGS cells or MKN45 cells, WB analysis (left) and densitometry analysis (right). n = 6 per group, *p < 0.05, **p < 0.01
Downregulated USP10 or TNFRSF10B promotes GC metastasis in vivo
In the metastatic tumor models, the MKN45 cells transfected with si-USP10, si-TNFRSF10B, or si-NC were intravenously injected into nude mice via the tail vein. After 5 weeks, the mice were euthanized, and lung metastatic nodules were observed in 3 out of 5 nude mice in each group. Compared with the si-NC group, the si-USP10 group or si-TNFRSF10B group had significantly more and larger lung metastatic nodules (Fig. 5). In addition, the AGS cells transfected with si-USP10 or si-NC were also intravenously injected into nude mice. Although 3 out of 5 nude mice in each group observed liver metastatic nodules instead of lung metastatic nodules, the number of liver metastatic nodules in the si-USP10 group was significantly higher than that in the si-NC group (Fig. S5). These data revealed that downregulated USP10 or TNFRSF10B promoted the metastatic ability of GC cells in vivo.
Downregulated USP10 or TNFRSF10B promotes GC lung metastasis in vivo. A Lung tissues were examined when mice were euthanized after MKN45 cells transfected with si-USP10 or si-TNFRSF10B were intravenously injected for 5 weeks (n = 5 per group, tumor formation rate = 60%), as well as the numbers of tumor nodules were counted and statistical analysis was performed. B The H&E staining in lung tissue sections. *p < 0.05
USP10 suppresses EMT via stabilizing TNFRSF10B expression
EMT can confer the migration and invasion ability of cancer cells [23], and we have previously reported that the expression of USP10 was positively correlated with the expression of E-cadherin in a large number of clinical samples [12]. Therefore, we detected the expression of E-cadherin, one of the markers of EMT, in AGS or MKN45 cells after USP10 knockdown or overexpression. In this study, we found that the protein expression levels of E-cadherin were downregulated in the group of si-USP10, and were upregulated in the group of GV657-USP10 (Fig. 6). The results demonstrated that downregulated USP10 induced EMT by regulating E-cadherin.
Figure 7A-C showed that the expressions of TNFRSF10B and E-cadherin were upregulated, while Twist1 expression was downregulated after GV657-USP10 transfected into AGS or MKN45 cells. Twist1 is one of the most important transcription factors, which negatively regulates E-cadherin [24, 25]. Thus, we speculated that USP10/TNFRSF10B may regulate the EMT of GC cells through the Twist1-E-cadherin pathway. The qRT-PCR assay demonstrated that the knockdown efficiency of si-TNFRSF10B-1/2/3 was very high, and si-TNFRSF10B-2 was used for follow-up experiments (Fig. 7D). E-cadherin expression was reduced, as well as Twist1 expression was increased after si-TNFRSF10B was transfected into AGS or MKN45 cells (Fig. 7E-G). Whereas, the mRNA expression level of Twist1 did not change significantly, indicating that TNFRSF10B and USP10 regulated Twist1 at the protein level (Fig. 7H). Furthermore, we found that USP10 overexpression after knock-down of TNFRSF10B in AGS or MKN45 cell lines, USP10 would be deprived of the ability for inhibiting Twist1 expression and upregulating E-cadherin expression (Fig. 7I-K). Thus, we believed that USP10 suppressed EMT through upregulating TNFRSF10B expression in GC cells.
USP10 suppressed EMT through stabilizing TNFRSF10B protein in GC cells. A, B and C The protein expression level of USP10, TNFRSF10B, Twist1, and E-cadherin were measured by WB after GV657-USP10 transfected into AGS cells or MKN45 cells. D The knockdown efficiency of si-TNFRSF10B-1, si-TNFRSF10B-2, or si-TNFRSF10B-3 was assessed by qRT-PCR. E, F and G The protein expression level of TNFRSF10B, Twist1, and E-cadherin were measured by WB after si-TNFRSF10B transfected into AGS cells or MKN45 cells. H The mRNA expression level of TNFRSF10B and Twist1 were measured by qRT-PCR after si-TNFRSF10B transfected into MKN45 cells. n = 6 per group, *p < 0.05, **p < 0.01. I, J and K The protein expression level of USP10, TNFRSF10B, Twist1, and E-cadherin were measured by WB after GV657-USP10 and si-TNFRSF10B transfected into AGS cells or MKN45 cells, n = 6 per group, *p < 0.05, **p < 0.01, vs. si-NC + GV657.
TNFRSF10B expression in human GC tissues
To investigate the correlation between TNFRSF10B mRNA expression and the prognosis of GAC patients, 433 samples in GSE84437 and 295 TCGA (STAD) samples were respectively used for survival analysis, and we used the Kaplan-Meier plotter (https://kmplot.com/analysis) including GEO, EGA, and TCGA databases to obtain more cohorts for survival analysis. Except for GSE51105, the GSE62254, GSE29272, GSE15459, and GSE84437 datasets analyses showed that TNFRSF10B mRNA expression was significantly positively correlated with the GAC patient overall survival (Fig. 8A and B). In addition, the results of the GSE62254 and GSE15459 datasets analysis demonstrated that the TNFRSF10B mRNA expression was significantly positively correlated with the first progression of GAC patients (Fig. S6A). Moreover, TNFRSF10B mRNA expression was significantly positively associated with the post progression survival of GAC patient in GSE62254 samples, while TNFRSF10B mRNA expression was not associated with the post progression survival of GAC patient in GSE15459 samples (Fig. S6B). However, TNFRSF10B mRNA expression was not associated with the overall survival of GAC patient in TCGA (STAD) samples (Fig. 8C). In our GAC cohort (135 males and 59 females; age range, 22–75 years old; mean age 59.7 ± 10.2 years old) study, we found that the TNFRSF10B protein expression was also not associated with the overall survival of GAC patient (Fig. 8D). Kaplan-Meier analysis showed that the decreased GAC patient survival was only significantly correlated with later TNM stage, one of the well-established prognostic factors (Fig. 8E), which demonstrated the representativeness of our GAC cohort. Next, the relationship between TNFRSF10B protein expression and the TNM stage of GAC was further analyzed in our 171 patients. The results showed that the expression rate of TNFRSF10B protein in the group without distant metastasis (M0) was significantly higher than that in the group with distant metastasis (M1), while no statistical difference was observed in T stage and N stage (Fig. 8F-H). Moreover, the expression trends between the T stage and the N stage or the M stage were opposite, which could partially explain why TNFRSF10B protein expression was not related to the survival of patients in our GAC cohort. Finally, we found that the expression rates of TNFRSF10B protein in the diffuse type GAC and poorly cohesive carcinoma were lower than that in the intestinal type GAC and non-poorly cohesive carcinoma, respectively (Fig. 8I and J), indicating that TNFRSF10B was involved in the regulation of GC cell adhesion. Representative histological features, IHC staining specimens, and scoring criteria were presented in Fig. 8K and L.
TNFRSF10B expression in human GC tissues. A The Kaplan-Meier overall survival analysis of TNFRSF10B in four GEO datasets of GC (https://kmplot.com/
analysis). B The relationship of TNFRSF10B mRNA expression to the overall survival of GAC patients in GSE84437 (p < 0.05). C TCGA datasets showed the relationship of TNFRSF10B mRNA expression to the overall survival of GAC patients (p > 0.05). D The relationship of TNFRSF10B protein expression to the overall survival of GAC patients in our cohort (p > 0.05). E The relationship of TNM stage to the overall survival of GAC patients in our cohort (p < 0.001). F The high expression rate of TNFRSF10B protein in depth of invasion T1, T2, T3, and T4 GAC (p > 0.05, Chi-square test for trend). G The high expression rate of TNFRSF10B protein in nodal metastasis N0 and N1/N2/N3 GAC (p > 0.05, Chi-square test for trend). H The high expression rate of TNFRSF10B protein in distant metastasis M0 and M1 GAC (p < 0.05, Chi-square test). I The high expression rate of TNFRSF10B protein in Lauren’s classification of intestinal type, mixed type, and diffuse type GAC (p < 0.001, Chi-square test). J The high expression rate of TNFRSF10B protein in poorly cohesive carcinoma and other adenocarcinoma GAC (p < 0.001, Chi-square test). K The H&E and IHC staining in the signet-ring cell carcinoma and tubular adenocarcinoma of paraffin-embedded GAC, green arrow: signet-ring cell carcinoma, red arrow: residual mucosal glands, original magnification, ×20, ×100. L The representative graph for immuno-staining score of TNFRSF10B, original magnification, ×200