Abstract
Background
Prostate Stem Cell Antigen (PSCA) is frequently expressed in prostate cancer but its exact function is unclear.
Methods
To clarify contradictory findings on the prognostic role of PSCA expression, a tissue microarray containing 13,665 prostate cancers was analyzed by immunohistochemistry.
Results
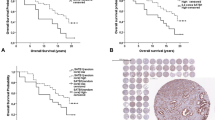
PSCA staining was absent in normal epithelial and stromal cells of the prostate. Membranous and cytoplasmic PSCA staining was seen in 53.7% of 9642 interpretable tumors. Staining was weak in 22.4%, moderate in 24.5% and strong in 6.8% of tumors. PSCA expression was associated with favorable pathological and clinical tumor features: Early pathological tumor stage (p < 0.0001), low Gleason grade (p < 0.0001), absence of lymph node metastasis (p < 0.0001), low pre-operative PSA level (p = 0.0118), negative surgical margin (p < 0.0001) and reduced PSA recurrence (p < 0.0001). PSCA expression was an independent predictor of prognosis in multivariate analysis (hazard ratio 0.84, p < 0.0001).
Conclusions
The absence of statistical relationship to TMPRSS2:ERG fusion status, chromosomal deletion or high tumor cell proliferation argues against a major role of PSCA for regulation of cell cycle or genomic integrity. PSCA expression is linked to favorable prognosis. PSCA measurement is a candidate for inclusion in multi-parametric prognostic prostate cancer tests.
Similar content being viewed by others
Background
While most prostate cancers have an indolent clinical course, the disease represents the third most common cause of cancer related death in men in Western societies [1]. Gleason grade and tumor extent on biopsies, preoperative prostate-specific antigen (PSA), and clinical stage are the currently established pretreatment prognostic parameters. Although these parameters are linked to cancer aggressiveness, the distinction between indolent and aggressive prostate cancer is difficult for the individual patient. Molecular marker may enable a better prediction of prostate cancer aggressiveness in the future.
Prostate stem cell antigen (PSCA) is a protein of unknown function anchored to the cell surface. It was discovered in an attempt to identify genes up regulated in human prostate cancer [2]. Though the name implies specificity for the prostate, PSCA is expressed in several tissues: Placenta, kidney, pancreas, and bladder [3,4,5]. The function of PSCA has not been fully elucidated [6,7,8,9]. Experiments suggest a possible role in cell adhesion, proliferation control and cell survival [2, 10]. Evidence is accumulating that – depending on the cell type involved – PSCA can have a tumor promoting or a tumor suppressive effect [11,12,13,14,15]. For example, loss of PSCA was associated with poor outcome in cancer of the gallbladder and stomach [12, 16], but with improved prognosis in pancreatic adenocarcinoma, renal cell carcinoma and non-small lung cancer [17,18,19,20]. The part of PSCA in prostate cancer remains unclear. Even if most available data suggest that prostate cancer may belong to the tumors with an oncogenic function of PSCA overexpression [46,47].
PSCA expression was significantly associated with favorable patient outcome in our cohort. A possible clinical relevance of this finding is supported by its statistical independence of classical prognostic markers, especially in a pre-operative disease state. However, in comparison with established features such as the Gleason score, the impact of PSCA expression on patient outcome was rather small. If traditional prognostic Gleason groups were used, a small prognostic impact was still found in Gleason 3 + 4 (p = 0.0206) or Gleason 4 + 3 (p = 0.0092). However, if these subgroups were further differentiated according to the fraction of Gleason 4 (quantitative Gleason grading [30]) these PSCA associated prognostic differences vanished. This further illustrates the high bar that molecular characteristics have to overcome if compared with optimized morphologic analysis.
Conclusions
PSCA expression is a statistically independent predictor of favorable prognosis in prostate cancer. Although its prognostic impact per se is not very strong, PSCA expression analysis could be considered for inclusion in multi-parametric prognostic tests to distinguish prostate cancers with need for radical therapy.
Abbreviations
- AR:
-
Androgen receptor
- CHD1:
-
Chromodomain-Helicase-DNA-Binding Protein 1
- ELAV1:
-
Embryonic Lethal, Abnormal Vision, Drosophila)-Like 1
- FISH:
-
Fluorescence in-situ hybridization
- FOXP1:
-
Forkhead box protein P1
- HOOK3:
-
Hook Microtubule Tethering Protein 3
- IHC:
-
Immunohistochemistry
- MAP3K:
-
Mitogen-Activated Protein Kinase Kinase Kinase 7
- PSCA:
-
Prostate stem cell antigen
- PSA:
-
Prostate specific antigen
- PTEN:
-
Phosphatase and tensin homolog
- RPE:
-
Radical prostatectomy
- TMPRSS2:
-
Transmembrane protease, serine 2
- ERG:
-
ETS-related gene fusion
- TMA:
-
Tissue microarray
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yamashiro J, Le Beau MM, et al. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 1998;95(4):1735–40.
Cunha AC, Weigle B, Kiessling A, Bachmann M, Rieber EP. Tissue-specificity of prostate specific antigens: comparative analysis of transcript levels in prostate and non-prostatic tissues. Cancer Lett. 2006;236(2):229–38.
Ross S, Spencer SD, Holcomb I, Tan C, Hongo J, Devaux B, Rangell L, Keller GA, Schow P, Steeves RM, et al. Prostate stem cell antigen as therapy target: tissue expression and in vivo efficacy of an immunoconjugate. Cancer Res. 2002;62(9):2546–53.
Gu Z, Thomas G, Yamashiro J, Shintaku IP, Dorey F, Raitano A, Witte ON, Said JW, Loda M, Reiter RE. Prostate stem cell antigen (PSCA) expression increases with high Gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000;19(10):1288–96.
Saeki N, Gu J, Yoshida T, Wu X. Prostate stem cell antigen: a Jekyll and Hyde molecule? Clin Cancer Res. 2010;16(14):3533–8.
Raff AB, Gray A, Kast WM. Prostate stem cell antigen: a prospective therapeutic and diagnostic target. Cancer Lett. 2009;277(2):126–32.
Esfahani M, Ataei N, Panjehpour M. Biomarkers for evaluation of prostate cancer prognosis. Asian Pac J Cancer Prev. 2015;16(7):2601–11.
Tricoli JV, Schoenfeldt M, Conley BA. Detection of prostate cancer and predicting progression: current and future diagnostic markers. Clin Cancer Res. 2004;10(12 Pt 1):3943–53.
Kim SH, Park WS, Kim SH, Park B, Joo J, Lee GK, Joung JY, Seo HK, Chung J, Lee KH. Prostate stem cell antigen expression in radical prostatectomy specimens predicts early biochemical recurrence in patients with high risk prostate Cancer receiving Neoadjuvant hormonal therapy. PLoS One. 2016;11(3):e0151646.
Zhang LY, Wu JL, Qiu HB, Dong SS, Zhu YH, Lee VH, Qin YR, Li Y, Chen J, Liu HB, et al. PSCA acts as a tumor suppressor by facilitating the nuclear translocation of RB1CC1 in esophageal squamous cell carcinoma. Carcinogenesis. 2016;37(3):320–32.
Ono H, Hiraoka N, Lee YS, Woo SM, Lee WJ, Choi IJ, Saito A, Yanagihara K, Kanai Y, Ohnami S, et al. Prostate stem cell antigen, a presumable organ-dependent tumor suppressor gene, is down-regulated in gallbladder carcinogenesis. Genes Chromosomes Cancer. 2012;51(1):30–41.
Saeki N, Ono H, Sakamoto H, Yoshida T. Down-regulation of immune-related genes by PSCA in gallbladder Cancer cells implanted into mice. Anticancer Res. 2015;35(5):2619–25.
Wang L, Sang Y, Tang J, Zhang RH, Luo D, Chen M, Deng WG, Kang T. Down-regulation of prostate stem cell antigen (PSCA) by slug promotes metastasis in nasopharyngeal carcinoma. J Pathol. 2015;237(4):411–22.
Zhigang Z, Wenlv S. Prostate stem cell antigen (PSCA) expression in human prostate cancer tissues: implications for prostate carcinogenesis and progression of prostate cancer. Jpn J Clin Oncol. 2004;34(7):414–9.
Study Group of Millennium Genome Project for C, Sakamoto H, Yoshimura K, Saeki N, Katai H, Shimoda T, Matsuno Y, Saito D, Sugimura H, Tanioka F, et al. genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40(6):730–40.
Amara N, Palapattu GS, Schrage M, Gu Z, Thomas GV, Dorey F, Said J, Reiter RE. Prostate stem cell antigen is overexpressed in human transitional cell carcinoma. Cancer Res. 2001;61(12):4660–5.
Argani P, Rosty C, Reiter RE, Wilentz RE, Murugesan SR, Leach SD, Ryu B, Skinner HG, Goggins M, Jaffee EM, et al. Discovery of new markers of cancer through serial analysis of gene expression: prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res. 2001;61(11):4320–4.
Elsamman EM, Fukumori T, Tanimoto S, Nakanishi R, Takahashi M, Toida K, Kanayama HO. The expression of prostate stem cell antigen in human clear cell renal cell carcinoma: a quantitative reverse transcriptase-polymerase chain reaction analysis. BJU Int. 2006;98(3):668–73.
Kawaguchi T, Sho M, Tojo T, Yamato I, Nomi T, Hotta K, Hamada K, Suzaki Y, Sugiura S, Kushibe K, et al. Clinical significance of prostate stem cell antigen expression in non-small cell lung cancer. Jpn J Clin Oncol. 2010;40(4):319–26.
Gu X, Ma Y, **ao J, Zheng H, Song C, Gong Y, **ng X. Up-regulated biglycan expression correlates with the malignancy in human colorectal cancers. Clin Exp Med. 2012;12(3):195–9.
Ruan Y, Yu W, Cheng F, Zhang X, Larre S. Detection of prostate stem cell antigen expression in human prostate cancer using quantum-dot-based technology. Sensors (Basel). 2012;12(5):5461–70.
Taeb J, Asgari M, Abolhasani M, Farajollahi MM, Madjd Z. Expression of prostate stem cell antigen (PSCA) in prostate cancer: a tissue microarray study of Iranian patients. Pathol Res Pract. 2014;210(1):18–23.
Han KR, Seligson DB, Liu X, Horvath S, Shintaku PI, Thomas GV, Said JW, Reiter RE. Prostate stem cell antigen expression is associated with Gleason score, seminal vesicle invasion and capsular invasion in prostate cancer. J Urol. 2004;171(3):1117–21.
Joung JY, Yang SO, Jeong IG, Han KS, Seo HK, Chung J, Park WS, Lee KH. Reverse transcriptase-polymerase chain reaction and immunohistochemical studies for detection of prostate stem cell antigen expression in prostate cancer: potential value in molecular staging of prostate cancer. Int J Urol. 2007;14(7):635–43.
Fuessel S, Sickert D, Meye A, Klenk U, Schmidt U, Schmitz M, Rost AK, Weigle B, Kiessling A, Wirth MP. Multiple tumor marker analyses (PSA, hK2, PSCA, trp-p8) in primary prostate cancers using quantitative RT-PCR. Int J Oncol. 2003;23(1):221–8.
Larkin SE, Holmes S, Cree IA, Walker T, Basketter V, Bickers B, Harris S, Garbis SD, Townsend PA, Aukim-Hastie C. Identification of markers of prostate cancer progression using candidate gene expression. Br J Cancer. 2012;106(1):157–65.
Moore ML, Teitell MA, Kim Y, Watabe T, Reiter RE, Witte ON, Dubey P. Deletion of PSCA increases metastasis of TRAMP-induced prostate tumors without altering primary tumor formation. Prostate. 2008;68(2):139–51.
Schmidt U, Fuessel S, Koch R, Baretton GB, Lohse A, Tomasetti S, Unversucht S, Froehner M, Wirth MP, Meye A. Quantitative multi-gene expression profiling of primary prostate cancer. Prostate. 2006;66(14):1521–34.
Sauter G, Steurer S, Clauditz TS, Krech T, Wittmer C, Lutz F, Lennartz M, Janssen T, Hakimi N, Simon R, et al. Clinical utility of quantitative Gleason grading in prostate biopsies and prostatectomy specimens. Eur Urol. 2016;69(4):592–8.
Mirlacher M, Simon R. Recipient block TMA technique. Methods Mol Biol. 2010;664:37–44.
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8.
Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, Simon R, Tennstedt P, Muller J, Scholz L, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17(18):5878–88.
Burkhardt L, Fuchs S, Krohn A, Masser S, Mader M, Kluth M, Bachmann F, Huland H, Steuber T, Graefen M, et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 2013;73(9):2795–805.
Kluth M, Hesse J, Heinl A, Krohn A, Steurer S, Sirma H, Simon R, Mayer PS, Schumacher U, Grupp K, et al. Genomic deletion of MAP3K7 at 6q12-22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod Pathol. 2013;26(7):975–83.
Krohn A, Seidel A, Burkhardt L, Bachmann F, Mader M, Grupp K, Eichenauer T, Becker A, Adam M, Graefen M, et al. Recurrent deletion of 3p13 targets multiple tumour suppressor genes and defines a distinct subgroup of aggressive ERG fusion-positive prostate cancers. J Pathol. 2013;231(1):130–41.
Minner S, Jessen B, Stiedenroth L, Burandt E, Kollermann J, Mirlacher M, Erbersdobler A, Eichelberg C, Fisch M, Brummendorf TH, et al. Low level HER2 overexpression is associated with rapid tumor cell proliferation and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16(5):1553–60.
Schlomm T, Iwers L, Kirstein P, Jessen B, Kollermann J, Minner S, Passow-Drolet A, Mirlacher M, Milde-Langosch K, Graefen M, et al. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol. 2008;21(11):1371–8.
Burdelski C, Dieckmann T, Heumann A, Hube-Magg C, Kluth M, Beyer B, Steuber T, Pompe R, Graefen M, Simon R, et al. p16 upregulation is linked to poor prognosis in ERG negative prostate cancer. Tumour Biol. 2016;37(9):12655–63.
Juhnke M, Heumann A, Chirico V, Hoflmayer D, Menz A, Hinsch A, Hube-Magg C, Kluth M, Lang DS, Moller-Koop C, et al. Apurinic/apyrimidinic endonuclease 1 (APE1/Ref-1) overexpression is an independent prognostic marker in prostate cancer without TMPRSS2:ERG fusion. Mol Carcinog. 2017;56(9):2135–45.
Wilczak W, Rashed S, Hube-Magg C, Kluth M, Simon R, Buscheck F, Clauditz TS, Grupp K, Minner S, Tsourlakis MC, et al. Up-regulation of mismatch repair genes MSH6, PMS2 and MLH1 parallels development of genetic instability and is linked to tumor aggressiveness and early PSA recurrence in prostate cancer. Carcinogenesis. 2017;38(1):19–27.
Melling N, Taskin B, Hube-Magg C, Kluth M, Minner S, Koop C, Grob T, Graefen M, Heinzer H, Tsourlakis MC, et al. Cytoplasmic accumulation of ELAVL1 is an independent predictor of biochemical recurrence associated with genomic instability in prostate cancer. Prostate. 2016;76(3):259–72.
Melling N, Harutyunyan L, Hube-Magg C, Kluth M, Simon R, Lebok P, Minner S, Tsourlakis MC, Koop C, Graefen M, et al. High-level HOOK3 expression is an independent predictor of poor prognosis associated with genomic instability in prostate Cancer. PLoS One. 2015;10(7):e0134614.
Brase JC, Johannes M, Mannsperger H, Falth M, Metzger J, Kacprzyk LA, Andrasiuk T, Gade S, Meister M, Sirma H, et al. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 2011;11:507.
Gupta S, Il** K, Sara H, Mpindi JP, Mirtti T, Vainio P, Rantala J, Alanen K, Nees M, Kallioniemi O. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70(17):6735–45.
Jhavar S, Brewer D, Edwards S, Kote-Jarai Z, Attard G, Clark J, Flohr P, Christmas T, Thompson A, Parker M, et al. Integration of ERG gene map** and gene-expression profiling identifies distinct categories of human prostate cancer. BJU Int. 2009;103(9):1256–69.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, **ao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22.
Acknowledgements
We thank Julia Schumann, Sünje Seekamp and Inge Brandt for excellent technical assistance.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its Additional file 1].
Author information
Authors and Affiliations
Contributions
MCH, AH, RS, and GS designed the study, and drafted the manuscript. HH1, MG, CB and TS participated in study design. MCH, AH, CG, CS1, CS2 performed IHC analysis and scoring. PL and EB participated in pathology data analysis. CH-M and RS performed statistical analysis. TS, CK, MK, MCH, HH2 and AH participated in data interpretation, and helped to draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The ethics committee of the Ärztekammer Hamburg approved this study (WF-049/09 and PV3652). According to local laws (HmbKHG, §12,1) informed consent was not required for this study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Figure S1. Kaplan-Meier plot of prostate specific antigen (PSA) recurrence and PSCA expression stratified for quantitative Gleason grade. Note the different time scale for Gleason Tertiary 5 grades. (DOC 2467 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Heinrich, MC., Göbel, C., Kluth, M. et al. PSCA expression is associated with favorable tumor features and reduced PSA recurrence in operated prostate cancer. BMC Cancer 18, 612 (2018). https://doi.org/10.1186/s12885-018-4547-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-018-4547-7