Abstract
Background
The purpose of the study is to discuss the correlation between the resistance rate of gram negative bacteria to fluoroquinolones (FQ) and antibiotic consumption intensity of 145 China tertiary hospitals in 2014.
Methods
This retrospective study adopted national monitoring data from 2014. Each participating hospital required to report annual consumption of each antibiotic, and the resistance rate of gram negative bacteria to FQ. Then the correlation between antibiotic usage and fluoroquinolones –resistant (FQR) rate was consequently investigated.
Results
One hundred forty-five hospitals were included in the study, and the median antibiotic consumption intensity was 46.30 (23.93–115.39) defined daily dosages (DDDs) per 100 patient-days. Cephalosporins ranks first in the antibiotics consumption, followed by fluoroquinolones, penicillins, and carbapenems. Fluoroquinolones resistance rate varied from hospital to hospital. The correlation analysis showed significant relationship between the percentage of FQR Escherichia coli and the consumption of FQs (r = 0.308, p<0.01) and levofloxacin (r = 0.252, p<0.01). For FQR Klebsiella pneumoniae, not only FQs (r = 0.291, p<0.01) and levofloxacin (r = 0.260, p<0.01) use but also carbapenems (r = 0.242, p<0.01) and overall antibiotics (r = 0.247, p<0.01) use showed significant correlation. The resistant proportion of FQR Pseudomonas aeruginosa was observed to be correlated with the consumption of all antibiotics (r = 0.260, p<0.01), FQs (r = 0.319, p<0.01) and levofloxacin (r = 0.377, p<0.01). The percentage of levofloxacin-resistant Acinetobacter baumannii was significantly correlated with the consumption of all antibiotics (r = 0.282, p<0.01), third-generation cephalosporins excluding combinations with beta-lactamase inhibitors (r = 0.246, p<0.01), FQs (r = 0.254, p<0.01) and levofloxacin (r = 0.336, p<0.01). However, the correlation of the ciprofloxacin-resistant A. baumannii and the antibiotics consumption was not found.
Conclusions
A strong correlation was demonstrated between the antibiotic consumption and the rates of FQR gram-negative bacteria. As unreasonable antibiotics usage remains crucial in the proceeding of resistant bacteria selection, our study could greatly promote the avoidance of unnecessary antibiotic usage.
Similar content being viewed by others
Background
Fluoroquinolones (FQs) were introduced as broad-spectrum antibiotics. FQs are widely used in infectious diseases due to their excellent oral bioavailability. With the widespread use of FQs, FQs-resistant (FQR) gram-negative bacteria are gradually increasing, limiting the selection for treating infections.
According to China antimicrobial resistance surveillance system in the first half of 2018, the ciprofloxacin-resistant rate was 57.8% for Escherichia coli, 35.4% for Klebsiella spp., 17.1% for Pseudomonas aeruginosa and 75.4% for Acinetobacter baumannii respectively. FQ resistance was reported to be an independent risk factor for hospital mortality, which have been associated with greater hospital expenses and poor clinical outcomes [1,2,3,4,5]. Moreover, compared with other phenotypic resistance patterns, bacteremia caused by FQR E. coli and Klebsiella spp. lead to higher mortality [6]. It is reported that previous colonization of FQR E. coli can lead to the spread of extended-spectrum beta-lactamase (ESBL) after the use of quinolone prophylaxis [5, 7]. The high probability of ESBL production by FQR gram-negative bacteria makes anti-infective treatment more difficult. Bloodstream infection caused by FQR gram-negative raised with the increased resistant rate to FQs, and the unadjusted mortality rate was 18% for FQR gram-negative bloodstream infections [1, 8].
It is plausible that the unreasonable antibiotics usage can induce the development of bacterial resistance. In this study, the aim was to investigat the correlation between resistance rate of gram-negative bacteria and antibiotic usage.
Methods
Study design
A cross-sectional study involving 145 voluntarily participating hospitals was conducted. Data on antibiotic consumption and the FQR rate of gram-negative bacteria from inpatients at each participating hospital in 2014 were collected. Then the antibiotic consumption and resistant rate was analyzed for possible correlation.
Data collection
Each participating hospital reported annual data of 2014, including antibiotic consumption and bacterial resistance data. The access to data acquisition can be found in the previous published paper, as well as the source of administrative data of involving hospital [9].
Measurement of antibiotic consumption and antibiotic resistance
Hospital pharmacists reported each antibiotic consumption to the national antibacterial drug clinical consumption survey network annually. According to Anatomic Therapeutic Chemical (ATC) classification system [10], data on consumption of all antibiotics (J01), beta-lactams (J01C + J01D), beta-lactam-beta-lactamase inhibitor combinations, beta-lactams excluding combinations with beta-lactamase inhibitors (CBLI), penicillins (J01C), penicillins excluding CBLI (J01C-J01CR), cephalosporins (J01DB + J01DC + J01DD + J01DE), cephalosporins excluding CBLI, third-generation cephalosporins (J01DD), third-generation cephalosporins excluding CBLI, fourth-generation cephalosporins (J01DE), carbapenems (J01DH), FQs (J01MA), ciprofloxacin, levofloxacin and moxifloxacin were analyzed. The measurement of antibiotics consumption intensity was the same as our previous published paper [9].
Each participating hospital was required to report information about E. coli, K. pneumoniae, P. aeruginosa and A. baumannii. The information collection of the admitted strains and quality control method of bacteria laboratory in each hospital were the same as our previous report [9]. All hospitals must adhere to the Clinical and Laboratory Standards Institute 2014 guidelines. The FQR rate was defined as the number of FQR isolates divided by the total isolates of the same species tested multiplied by 100. FQs-resistance was defined as a strain resistant to levofloxacin or ciprofloxacin. The FQR rate was considered as the higher resistance rate if the resistance rates were different. For A. baumannii, ciprofloxacin and levofloxacin-resistant rate were derived separately because of their great differences. All antibiotic resistance data extraction were performed by the operation of Whonet 5.6 software.
Statistical analysis
With pearson’s correlation performance, the correlation between annual antibiotic consumption and FQR rate of gram-negative bacteria from 145 hospitals of China was conducted. Through the conduction of Microsoft Excel 2013 and STATA 16 (StataCorp LLC, Texas, USA), the correlation analysis was realized between the FQR rate of gram-negative bacteria and the antibiotic consumption intensity. Statistical significance was defined as p < 0.05.
Results
Participating hospitals
Altogether 145 hospitals were included in this study, 141 of the participating hospitals were tertiary hospitals. There are 29 hospitals in North China, 29 hospitals in East China, 22 hospitals in Central China, 25 hospitals in Southern China, 12 hospitals in the southwest of China, 15 hospitals in the northwest of China, and 13 hospitals in northeast of China. These hospitals had a median of 2356 beds (range, 720–8475 beds). The median number of inpatients per year reached 88.3 thousands (range, 10–410 thousands).
Antibiotics consumption
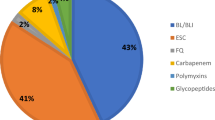
The median of the overall antibiotics consumption intensity in the included hospital was 46.30 DDDs/100 patient-days (23.93–115.39 DDDs/100 patient-days). Among them, cephalosporins were the most- prescribed antibiotics, followed by fluoroquinolones, penicillins and carbapenems. The antibiotics consumption intensity of the main antibiotic classes was depicted in Table 1.
Correlation between antibiotics consumption intensity and FQR E. coli
One hundred thirty-eight hospitals were admitted to perform the correlation between antibiotics consumption intensity and rate of FQR E. coli. One hundred fifty-eight thousand eight hundred sixty-six strains of E. coli were isolated, and 94,247 strains of them were FQR. The median resistance rate of E. coli to FQ was 61.67% (44.96–82.2%). As shown in Table 2 and Fig. 1, the resistance rate of E. coli to FQ was positively correlated with the consumption of FQs (r = 0.308, p<0.01) and levofloxacin (r = 0.252, p<0.01).
Correlation between antibiotics consumption intensity and FQR K. pneumoniae
One hundred thirty-nine hospitals were admitted to analyze the correlation between antibiotic consumption intensity and rate of FQR K. pneumoniae. One hundred ninety-four thousand nine hundred fifty-seven strains of K. pneumoniae were isolated, and 48,287 strains of them were FQR. The median resistance rate of K. pneumoniae to FQ was 23.1% (5.3–66.9%). As depicted in Table 2 and Fig. 2, the resistance rate of K. pneumoniae to FQ was positively correlated with the consumption of all antibiotics (r = 0.247, p<0.01), carbapenems (r = 0.242, p<0.01), FQ (r = 0.291, p<0.01) and levofloxacin (r = 0.260, p<0.01).
Correlation between antibiotics consumption intensity and FQR P. aeruginosa
One hundred thirty-nine hospitals were admitted to determine the correlation between antibiotic consumption intensity and rate of FQR P. aeruginosa. One hundred eleven thousand seven hundred eleven strains of P. aeruginosa were isolated, and 27,171 strains of them were FQR. The median resistance rate of P. aeruginosa to FQ was 22.4% (7.7–65.2%). As demonstrated in Table 2 and Fig. 3, the resistance rate of P. aeruginosa to FQ was positively correlated with the consumption of all antibiotics (r = 0.260, p<0.01), FQs (r = 0.319, p<0.01) and levofloxacin (r = 0.377, p<0.01).
Correlation between antibiotics consumption intensity and FQR A. baumannii
One hundred thirty-one hospitals were admitted to conduct the correlation between antibiotic consumption intensity and rate of levofloxacin-resistant A. baumannii. Ninety-three thousand one hundred fourteen strains of A. baumannii were isolated, and 52,695 strains of them were levofloxacin-resistant. The median resistance rate of A. baumannii to levofloxacin was 59.3% (16.1–93.9%). As suggested in Table 2 and Fig. 4, resistance rate of A. baumannii to FQ was positively correlated with the consumption of all antibiotics (r = 0.282, p<0.01), third-generation cephalosporins excluding CBLI (r = 0.246, p<0.01), FQs (r = 0.254, p<0.01) and levofloxacin (r = 0.336, p<0.01)..
One hundred thirty-three hospitals were admitted to analyze the correlation between antibiotic consumption intensity and rate of ciprofloxacin-resistant A. baumannii. One hundred one thousand three hundred seventy-four strains of A. baumannii were isolated, and 80,032 strains of them were ciprofloxacin-resistant. The median resistance rate of A. baumannii to ciprofloxacin was 77.6% (28.7–95%). However, as shown in Table 2, the percentage of ciprofloxacin-resistant A. baumannii was not significantly correlated with the antibiotics consumption..
Discussion
This study reflected the current status of antibiotic usage and gram-negative bacilli of fluoroquinolones-resistant patterns at the hospital level in China. During the study period, cephalosporins were the most- prescribed antibiotics, followed byfluoroquinolone, penicillins and carbapenems. Our data demonstrated that the percentage of FQR E. coli was significantly correlated with the consumption of FQs and levofloxacin while the rate of FQR K. pneumoniae was significantly correlated with the consumption of all antibiotics, carbapenems, FQs and levofloxacin. When it comes to FQR P. aeruginosa, the resistant rate was significantly correlated with the consumption of all antibiotics, FQs and levofloxacin. Furthermore, the percentage of levofloxacin-resistant A. baumannii was significantly correlated with the consumption of all antibiotics, third-generation cephalosporins excluding combinations with beta-lactamase inhibitors, FQs and levofloxacin. However, the correlation of ciprofloxacin-resistant A. baumannii and the antibiotics consumption was not found.
Consistant with previous studies, our results found that the FQR E. coli was significantly associated with FQs consumption [10,11,12,13,14,15,16,17,18,19]. The most important resistance mechanism of E. coli to quinolones is mutation of target gene DNA gyrase and topoisomerase IV. Further, up expression of active efflux pump, changes in membrane permeability of bacteria and plasmid-mediated quinolone resistance are the main mechanism of FQR E. coli production [20]. However, the emergence of FQR E. coli occurs as a multistep process, with increasing numbers of target gene mutations leading to progressively higher minimum inhibitory concentration s[21]. Not surprisingly, the most important risk factor for FQR E. coli appears to be previous FQs use [22,23,24]. Restriction of quinolone use can offer opportunities to reduce the prevalence of FQR E. coli [25,26,27].
However, data from 42 Spanish hospitals collected by the European Antimicrobial Surveillance Network indicated that amoxicillin/clavulanic acid use was the main driving force for the progression of FQR E. coli, possibly due to its high consumption in Spain [13]. Thirty-six acute care hospitals from France indicated the level of first, second and third-generation cephalosporins, as well as tetracycline’s usage influenced the incidence of FQR E. coli [15]. Compared with the third-generation cephalosporins, the resistance rate of E.coli to fluoroquinolones is higher, so the third-generation cephalosporins will be extensively used when the infection of E.coli is suspected. The author believe that the reason for the correlation between FQR E. coli and tegacycline use is that FQR E. coli is plausibly associated with resistance to tetracyclines.
There is mounting evidence demonstrating that the prevalence of ciprofloxacin-resistant K. pneumoniae was associated with use of ciprofloxacin and FQs [18, 27, 28]. A study from database of the Korean Health Insurance Review and Assessment Service suggested that the consumption of all third-generation cephalosporins was significantly correlated with resistance rates of K. pneumoniae to levofloxacin with a quarter lag [29]. The resistance mechanism of K. pneumoniae to FQs is the change of target sites, the change of outer membrane protein permeability, the effect of efflux pump and the transfer of resistant plasmids among bacteria. Our study results suggested that the percentage of FQR K. pneumoniae was significantly correlated with the consumption of all antibiotics, carbapenems, FQs and levofloxacin. The use of FQs is a risk factor for the incidence of FQR K. pneumoniae [30]. Therefore, it is no doubt that the overuse of FQs increases FQR K. pneumoniae.
The drug resistance of K. pneumoniae is mostly mediated by plasmids. Plasmid DNA can carry multiple drug-resistant genes such as qnr, ESBLs, Amp C enzyme and metalloenzyme-coded genes at the same time. As a mobile genetic primitive, plasmids can transmit carbapenemase-producing resistance genes, resulting in the outbreaks of multidrug-resistant K. pneumoniae [31]. Therefore, it is credible that the production of multidrug-resistant K. pneumoniae was correlated with the use of carbapenems and all antibiotics.
Resistance to fluoroquinolones developed in P. aeruginosa by various mechanisms: mutations in the genes encoding bacterial DNA topoisomerase II and topoisomerase IV is a major cause of resistance to fluoroquinolones in P. aeruginosa isolates. Meanwhile, overexpression of active efflux systems can reduce the permeability of the membrane. Furthermore,it is reported that plasmid carrying ESBL gene can also simultaneously carry quinolones resistance gene, which may be the reason of high level of resistance to quinolones in ESBL-producing strains [32].
Consistent with our study, the positive correlation between FQs consumption and the rates of FQR P. aeruginosa was described [18, 27, 33,34,35]. Moreover, the increased consumption of levofloxacin was explored to have correlation with raised resistance rate of P. aeruginosa to FQ but not ciprofloxacin [36,37,38,39,40]. Compared with ciprofloxacin, levofloxacin has weaker anti- P. aeruginosa activity, and it is easier to screen out the colonization or infection of FQR P. aeruginosa. However, various studies indicated that the consumption of ceftazidime, anti-pseudomonal cephalosporin and ciprofloxacin was positively correlated with the incidence rates of ciprofloxacin-resistant P. aeruginosa [35, 38, 41]. As we known, the use of certain antibiotics could be both a cause and a consequence of the resistance emergence. So it seems reasonable to explain high consumption of ceftazidime, anti-pseudomonal cephalosporin and ciprofloxacin in hospitals is likely to be a result of the high prevalence of FQR organisms and the alternative drugs usage to treat infection.
A. baumannii has become a difficult bacteria in clinical treatment because of its complex drug resistance mechanism and high drug resistance rate. The use of wide spectrum antibiotics will further screen out multidrug-resistant bacteria. Our study showed that the resistance rate of A. baumannii to ciprofloxacin was higher than that of levofloxacin, probably because ciprofloxacin is easier to detect the overexpression of pumps [42]. Our study illustrated the selection pressure of FQs use in the development of FQR A. baumannii, which is in accordance with a nationwide multicenter study from Kore a[18]. The drug resistance mechanism of A. baumannii to quinolones modified the bacterial DNA helix enzyme through the mutation of quinolone resistance gene cluster, thus reducing the affinity between the drug and the enzyme-DNA complex and leading to drug resistance. Also, some efflux systems affect the drug sensitivity. Up to now, three types of plasmid-mediated quinolone resistance genes have been identified, namely aminoglycoside acetyltransferase AAC (6′) - Ib-cr, specific efflux systems Qep A and Oqx AB and Qnr family [43]. Furthermore, simultaneous mutations in gyrA and parC genes have great influence on high-level fluoroquinolone resistance in A. baumannii [44].
Amp C enzyme is a cephalosporin enzyme encoded by chromosomes inherent in all A. baumannii. Adding the promoter insertion sequence ISAba1 beside the Amp C gene increases the production of beta-lactamase, which leads to resistance to cephalosporins. This also explains the correlation between drug resistance and consumption of 3GC excluding CBLI [45]. A domestic research report that the consumption of carbapenem has a significant positive relationship with A. baumannii resistance to levofloxacin [46]. A retrospective study manifested the consumption of cefmetazole and total cephamycin positively correlated with the resistance rates of A. baumannii to levofloxacin. The author believed that these consequences may be partly due to production of AmpC enzymes [47].
With the extensive use of FQs, their adverse reactions and the harm caused by irrational use have become increasingly prominent in recent years. Therefore, drug regulatory agencies at home and abroad frequently issued drug safety warnings, demanding the discontinuation or restriction of the FQs use. In our survey, the resistant rates of all gram-negative bacteria were significantly correlated with the FQs consumption, which is consistant with previous study. It is more likely that the resistance mechanism of gram-negative bacteria to FQs leads to a correlation between resistance and FQs consumption. Therefore, in view of the adverse reactions of FQs and the increasing drug resistance, controlling FQs consumption should be taken seriously.
Antibiotic use is explored to have correlation with the development of antibiotic resistance but the relationship is a multifaceted issue such as cross-transmission, interhospital transfer of resistance, a community contribution to resistance and so on. In our study, the correlation analysis between FQs use and FQ-resistant rate was based on hospital-level perspective, not that of the patient. Therefore, the results may be subject to ecological bias, suggesting that this study dose not reflect patient-level relationships. Additionally, this study did not include the use of outpatient antibiotics, which would underestimate the magnitude of association between FQs exposure and resistant rate to FQ. At the same time, if we can know the phenotype of the resistant isolates, we can definitely know whether any outbreak of cloned strains has affected our research results. If clonal dissemination of resistant strains contributed in part of the resistant organisms present, then a weaker association between antibiotic exposure and resistance might be found. While we acknowledge that our research did not take chronology into account, which is an inevitable defect of the design. This may also be the reason for the not very high correlation coefficient. Even so, it must be stated that the scale of the study made it the most comprehensive investigation reported domestically.
Conclusion
To summarize, our study quantified the associations between antibiotic consumption and the incidence of FQR gram-negative bacteria. As prudent and appropriate use of antibiotics plays a vital role in preventing the selection of resistant bacteria, our study results could serve as a driving force for implementation of antimicrobial stewardship policies.
Availability of data and materials
The data used in the current study are available from the corresponding author on reasonable request.
Abbreviations
- A.baumannii :
-
Acinetobacter baumannii
- ATC:
-
Anatomic Therapeutic Chemical
- DDDs:
-
Defined daily dosages
- E. coli :
-
Escherichia coli
- ESBLs:
-
Extended-spectrum beta-lactamase
- FQs:
-
Fluoroquinolones
- FQR:
-
fluoroquinolones-resistant
- K. pneumoniae :
-
Klebsiella pneumoniae
- P. aeruginosa :
-
Pseudomonas aeruginosa
References
Kadri SS, Adjemian J, Lai YL, Spaulding AB, Ricotta E, Prevots DR, et al. Difficult-to-Treat Resistance in Gram-negative Bacteremia at 173 US Hospitals: Retrospective Cohort Analysis of Prevalence, Predictors, and Outcome of Resistance to All First-line Agents. Clin Infect Dis. 2018;67:1803–14.
Camins BC, Marschall J, DeVader SR, Maker DE, Hoffman MW, Fraser VJ. The clinical impact of fluoroquinolone resistance in patients with E coli bacteremia. J Hosp Med. 2011;6:344–9.
Lautenbach E, Metlay JP, Bilker WB, Edelstein PH, Fishman NO. Association between fluoroquinolone resistance and mortality in Escherichia coli and Klebsiella pneumoniae infections: the role of inadequate empirical antimicrobial therapy. Clin Infect Dis. 2005;41:923–9.
Gasink LB, Fishman NO, Weiner MG, Nachamkin I, Bilker WB, Lautenbach E. Fluoroquinolone-resistant Pseudomonas aeruginosa: assessment of risk factors and clinical impact. Am J Med. 2006;119:526.e19–25.
Sawa T, Shimizu M, Moriyama K, Wiener-Kronish JP. Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: a review. Crit Care. 2014;18:668.
Suzuki H, Perencevich EN, Livorsi DJ, Alexander B, Beck BF, Richardson KK, et al. Attributable mortality due to fluoroquinolone and extended-spectrum cephalosporin resistance in hospital-onset Escherichia coli and Klebsiella spp. bacteremia: A matched cohort study in 129 Veterans Health Administration medical centers. Infect Control Hosp Epidemiol. 2019;40:928–31.
Chong Y, Shimoda S, Yakushiji H, Ito Y, Aoki T, Miyamoto T, et al. Clinical impact of fluoroquinolone-resistant Escherichia coli in the fecal flora of hematological patients with neutropenia and levofloxacin prophylaxis. PLoS One. 2014;9:e85210.
Blennow O, Ljungman P, Sparrelid E, Mattsson J, Remberger M. Incidence, risk factors, and outcome of bloodstream infections during the pre-engraftment phase in 521 allogeneic hematopoietic stem cell transplantations. Transpl Infect Dis. 2014;16:106–14.
Yang P, Chen Y, Jiang S, Shen P, Lu X, **ao Y. Association between antibiotic consumption and the rate of carbapenem-resistant Gram-negative bacteria from China based on 153 tertiary hospitals data in 2014. Antimicrob Resist Infect Control. 2018;19:137.
McDonnell L, Armstrong D, Ashworth M, Dregan A, Malik U, White P. National disparities in the relationship between antimicrobial resistance and antimicrobial consumption in Europe: an observational study in 29 countries-authors’ response. J Antimicrob Chemother. 2017;72:3500.
Gallini A, Degris E, Desplas M, Bourrel R, Archambaud M, Montastruc JL, et al. Influence of fluoroquinolone consumption in inpatients and outpatients on ciprofloxacin-resistant Escherichia coli in a university hospital. J Antimicrob Chemother. 2010;65:2650–7.
Hsu LY, Tan TY, Tam VH, Kwa A, Fisher DA, Koh TH, et al. Surveillance and correlation of antibiotic prescription and resistance of Gram-negative bacteria in Singaporean hospitals. Antimicrob Agents Chemother. 2010;54:1173–8.
Cuevas O, Oteo J, Lázaro E, Aracil B, de Abajo F, García-Cobos S, et al. Significant ecological impact on the progression of fluoroquinolone resistance in Escherichia coli with increased community use of moxifloxacin, levofloxacin and amoxicillin/clavulanic acid. J Antimicrob Chemother. 2011;66:664–9.
Wu HH, Liu HY, Lin YC, Hsueh PR, Lee YJ. Correlation between levofloxacin consumption and the incidence of nosocomial infections due to fluoroquinolone-resistant Escherichia coli. J Microbiol Immunol Infect. 2016;49:424–9.
Batard E, Ollivier F, Boutoille D, Hardouin JB, Montassier E, Caillon J, et al. Relationship between hospital antibiotic use and quinolone resistance in Escherichia coli. Int J Infect Dis. 2013;17:e254–8.
Hsueh PR, Chen WH, Luh KT. Relationships between antimicrobial use and antimicrobial resistance in Gram-negative bacteria causing nosocomial infections from 1991 to 2003 at a university hospital in Taiwan. Int J Antimicrob Agents. 2005;26:463–72.
Kim YA, Park YS, Youk T, Lee H, Lee K. Trends in South Korean antimicrobial use and association with changes in Escherichia coli resistance rates: 12-year ecological study using a nationwide surveillance and antimicrobial prescription database. PLoS One. 2018;13:e0209580.
Kim B, Kim Y, Hwang H, Kim J, Kim SW, Bae IG, et al. Trends and correlation between antibiotic usage and resistance pattern among hospitalized patients at university hospitals in Korea, 2004 to 2012: A nationwide multicenter study. Medicine (Baltimore). 2018;97:e13719.
Cusini A, Herren D, Bütikofer L, Plüss-Suard C, Kronenberg A, Marschall J. Intra-hospital differences in antibiotic use correlate with antimicrobial resistance rate in Escherichia coli and Klebsiella pneumoniae: a retrospective observational study. Antimicrob Resist Infect Control. 2018;7.
Redgrave LS, Sutton SB, Webber MA, Piddock LJ. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014;22:438–45.
Singh R, Swick MC, Ledesma KR, Yang Z, Hu M, Zechiedrich L, et al. Temporal interplay between efflux pumps and target mutations in development of antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother. 2012;56:1680–5.
Bedoin M, Cazorla C, Lucht F, Berthelot P, Boyer M, Carricajo A, et al. Risk factors for quinolone-resistance in women presenting with Escherichia coli acute pyelonephritis. Med Mal Infect. 2014;44:206–16.
Kim J, Kang CI, Joo EJ, Ha YE, Cho SY, Gwak GY, et al. Risk factor of community-onset spontaneous bacterial peritonitis caused by fluoroquinolone-resistant Escherichia coli in patients with cirrhosis. Liver Int. 2014;34:695–9.
Katsandri A, Avlamis A, Vasilakopoulou A, Mela V, Kosmidis C, Papaparaskevas J, et al. Risk factors for coexistence of fluoroquinolone resistance and ESBL production among Enterobacteriaceae in a Greek university hospital. J Chemother. 2008;20:452–7.
Gottesman BS, Carmeli Y, Shitrit P, Chowers M. Impact of quinolone restriction on resistance patterns of Escherichia coli isolated from urine by culture in a community setting. Clin Infect Dis. 2009;49:869–75.
Sarma JB, Marshall B, Cleeve V, Tate D, Oswald T, Woolfrey S. Effects of fluoroquinolone restriction (from 2007 to 2012) on resistance in Enterobacteriaceae: interrupted time-series analysis. J Hosp Infect. 2015;91:68–73.
Wang H, Wang H, Yu X, Zhou H, Li B, Chen G, et al. Impact of antimicrobial stewardship managed by clinical pharmacists on antibiotic use and drug resistance in a Chinese hospital, 2010–2016: a retrospective observational study. BMJ Open. 2019;9:e026072.
Lee HS, Loh YX, Lee JJ, Liu CS, Chu C. Antimicrobial consumption and resistance in five Gram-negative bacterial species in a hospital from 2003 to 2011. J Microbiol Immunol Infect. 2015;48:647–54.
Ryu S, Klein EY, Chun BC. Temporal association between antibiotic use and resistance in Klebsiella pneumoniae at a tertiary care hospital. Antimicrob Resist Infect Control. 2018;7:83.
Bolon MK, Wright SB, Gold HS, Carmeli Y. The magnitude of the association between fluoroquinolone use and quinolone-resistant Escherichia coli and Klebsiella pneumoniae may be lower than previously reported. Antimicrob Agents Chemother. 2004;48:1934–40.
Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–75.
Subedi D, Vijay AK, Willcox M. Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: an ocular perspective. Clin Exp Optom. 2018;101:162–71.
Lafaurie M, Porcher R, Donay JL, Touratier S, Molina JM. Reduction of fluoroquinolone use is associated with a decrease in methicillin-resistant Staphylococcus aureus and fluoroquinolone-resistant Pseudomonas aeruginosa isolation rates: a 10 year study. J Antimicrob Chemother. 2012;67:1010–5.
Weng TC, Chen YH, Lee CC, Wang CY, Lai CC, Tang HJ, et al. Correlation between fluoroquinolone consumption in hospitals and ciprofloxacin resistance amongst Pseudomonas aeruginosa isolates causing healthcare-associated infections, Taiwan, 2000–2009. Int J Antimicrob Agents. 2011;37:581–4.
Rogues AM, Dumartin C, Amadéo B, Venier AG, Marty N, Parneix P, et al. Relationship between rates of antimicrobial consumption and the incidence of antimicrobial resistance in Staphylococcus aureus and Pseudomonas aeruginosa isolates from 47 French hospitals. Infect Control Hosp Epidemiol. 2007;28:1389–95.
Lee YJ, Liu HY, Lin YC, Sun KL, Chun CL, Hsueh PR. Fluoroquinolone resistance of Pseudomonas aeruginosa isolates causing nosocomial infection is correlated with levofloxacin but not ciprofloxacin use. Int J Antimicrob Agents. 2010;35:261–4.
Kaye KS, Kanafani ZA, Dodds AE, Engemann JJ, Weber SG, Carmeli Y. Differential effects of levofloxacin and ciprofloxacin on the risk for isolation of quinolone-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2006;50:2192–6.
Mohr JF, Jones A, Ostrosky-Zeichner L, Wanger A, Tillotson G. Associations between antibiotic use and changes in susceptibility patterns of Pseudomonas aeruginosa in a private, university-affiliated teaching hospital: an 8-year-experience: 1995–2002. Int J Antimicrob Agents. 2004;24:346–51.
Pakyz AL, Lee JA, Ababneh MA, Harpe SE, Oinonen MJ, Polk RE. Fluoroquinolone use and fluoroquinolone-resistant Pseudomonas aeruginosa is declining in US academic medical centre hospitals. J Antimicrob Chemother. 2012;67:1562–4.
Polk RE, Johnson CK, McClish D, Wenzel RP, Edmond MB. Predicting hospital rates of fluoroquinolone-resistant Pseudomonas aeruginosa from fluoroquinolone use in US hospitals and their surrounding communities. Clin Infect Dis. 2004;39:497–503.
Gbaguidi-Haore H, Dumartin C, L’Hériteau F, Péfau M, Hocquet D, Rogues AM, et al. Antibiotics involved in the occurrence of antibiotic-resistant bacteria: a nationwide multilevel study suggests differences within antibiotic classes. J Antimicrob Chemother. 2013;68:461–70.
Güler G, Eraç B. Investigation of fluoroquinolone resistance mechanisms in clinical Acinetobacter baumannii isolates. Mikrobiyol Bul. 2016;50:278–86.
Asif M, Alvi IA, Rehman SU. Insight into Acinetobacter baumannii: pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect Drug Resist. 2018;11:1249–60.
Ardebili A, Lari AR, Beheshti M, Lari ER. Association between mutations in gyrA and parC genes of Acinetobacter baumannii clinical isolates and ciprofloxacin resistance. Iran J Basic Med Sci. 2015;18:623–6.
Hamidian M, Hall RM. Resistance to third-generation cephalosporins in Acinetobacter baumannii due to horizontal transfer of a chromosomal segment containing ISAba1-ampC. J Antimicrob Chemother. 2014;69:2865–6.
Cao J, Song W, Gu B, Mei YN, Tang JP, Meng L, et al. Correlation between carbapenem consumption and antimicrobial resistance rates of Acinetobacter baumannii in a university-affiliated hospital in China. J Clin Pharmacol. 2013;53:96–102.
Song W, Cao J, Mei YL. Correlation between cephamycin consumption and the incidence of antimicrobial resistance in Acinetobacter baumannii at a university hospital in China from 2001 to 2009. Int J Clin Pharmacol Ther. 2011;49:765–71.
Acknowledgements
Not applicable.
Funding
This study was supported by National Natural Science Foundation of China (81361138021, 81711530049 and 81800594), Key Research and Development Program of Zhejiang Province (2015C03032). The funders were not involved in the design of the study, data collection, analysis and interpretation of the data and the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
PY was responsible for the correlation analysis and manuscript writing. YC and PS conducted the bacterial resistance data analysis. SJ and XL analyzed antibiotic consumption intensity. XH as a principal investigator, contributed in the design of the study, results interpretation and manuscript review. The final manuscript was approved by all authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Approval of the ethics committee was not required in this study.
Consent for publication
Not applicable.
Competing interests
The authors report no conflicts of interest relevant to this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yang, P., Chen, Y., Jiang, S. et al. Association between the rate of fluoroquinolones-resistant gram-negative bacteria and antibiotic consumption from China based on 145 tertiary hospitals data in 2014. BMC Infect Dis 20, 269 (2020). https://doi.org/10.1186/s12879-020-04981-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-020-04981-0