Abstract
Background
Abscisic acid (ABA) plays a crucial role in abiotic stress responses. The pyrabactin resistance (PYR)/PYR-like (PYL)/regulatory component of ABA receptor (RCAR) proteins that have been characterized as ABA receptors function as the core components in ABA signaling pathway. However, the functions of grape PYL genes in response to different abiotic stresses, particularly cold stress, remain less studied.
Results
In this study, we investigated the expression profiles of grape PYL genes upon cold treatment and isolated the VaPYL4 gene from Vitis amurensis, a cold-hardy grape species. Overexpression of VaPYL4 gene in grape calli and Arabidopsis resulted in enhanced cold tolerance. Moreover, plant resistance to drought and salt stress was also improved by overexpressing VaPYL4 in Arabidopsis. More importantly, we evaluated the contribution of VaPYL4 to plant growth and development after the treatment with cold, salt and drought stress simultaneously. The transgenic plants showed higher survival rates, earlier flowering phenotype, and heavier fresh weight of seedlings and siliques when compared with wild-type plants. Physiological analyses showed that transgenic plants had much lower content of malondialdehyde (MDA) and higher peroxidase (POD) activity. Stress-responsive genes such as RD29A (Responsive to desiccation 29A), COR15A (Cold responsive 15A) and KIN2 (Kinase 2) were also significantly up-regulated in VaPYL4-overexpressing Arabidopsis plants.
Conclusions
Our results show that overexpression of VaPYL4 could improve plant performance upon different abiotic stresses, which therefore provides a useful strategy for engineering future crops to deal with adverse environments.
Similar content being viewed by others
Background
Plant growth and development are usually challenged by a variety of abiotic stresses such as cold, drought and salt stress. These abiotic stresses are big concerns for agriculture and could result in the loss of crop productivity [1]. After exposure to abiotic stress, plants need to coordinate physiological and biochemical processes and also gene expression to adapt to the severe environmental conditions [2]. Gene expression is generally associated with stress-induced hormone signaling, and phytohormone abscisic acid (ABA) has been reported to play a vital role in plant response to abiotic stress [3,4,5,6].
ABA signaling is initially triggered by ABA perception, which is accomplished by the binding of ABA receptors to ABA [7]. The pyrabactin resistance (PYR)/PYR-like (PYL)/regulatory component of ABA receptor (RCAR) that localizes to nucleus and cytosol is the predominant type of ABA receptors [7]. The PYR/PYL/RCAR (hereafter referred to as PYL) ABA receptor, together with protein phosphatase 2C (PP2C) and SNF1-related protein kinase 2 (SnRK2), has been reveled to form the core ABA signaling network, which is characterized as double-negative regulatory system [7, 8]. In the absence of ABA, PP2Cs bind and dephosphorylate SnRK2s, inhibiting the activities of SnRK2 proteins. When ABA molecules are recognized and bound by ABA receptors, the ABA-receptor complexes could physically interact with PP2Cs, resulting in the release of SnRK2s, which can activate the expression of downstream target genes [8,9,10,11]. In plants, many ABA receptors, protein phosphatases and kinases have been identified as ABA signaling components. In Arabidopsis thaliana, for example, there are 14 PYL members, 76 members of PP2C proteins and 10 SnRK2 protein kinases [12,13,14]. Following the study in Arabidopsis, the members of PYL, PP2C and SnRK2 family have also been isolated in other plants, such as rice [13, 15], maize [16,17,18], and tomato [19]. The ABA signal transduction pathway has been characterized in grapevine (Vitis vinifera) [20], and the members of grape PYL, PP2C and SnRK2 family are 9, 85 and 7, respectively [21].
As the stress phytohormone, ABA accumulation is rapidly increased in plants after exposure to abiotic stress, particularly drought and salinity [3, 8, 22]. The functions of ABA receptors in response to abiotic stress have been revealed in plants these years [23, 24]. Overexpression of AtPYL4/RCAR10, AtPYL5/RCAR8 and AtPYL13/RCAR10 enhanced drought resistance in transgenic Arabidopsis [25,26,27]. The AtPYL9/RCAR1 was found to promote leaf senescence, which in turn increases drought resistance by limiting transpirational water loss and promoting water to flow to young tissues [27]. Moreover, Arabidopsis ABA receptor genes AtPYL1/RCAR12 and AtPYL3/RCAR13 were found to play a role in response to extreme temperatures, and their overexpressing plants showed increased tolerance to both cold and heat stress [28]. Ectopic overexpression of OsPYL3 in Arabidopsis led to enhanced tolerance to drought and cold stress [29]. Similar results were also obtained when using the OsPYL10 gene, whose overexpression resulted in improved tolerance to drought and cold stress in transgenic rice [30]. In 2012, VvPYL1 was first identified as an ABA receptor in grape [31]. Later, potential ABA receptors were systematically characterized in grape, and VvRCAR7 was revealed to be induced by drought, salt and cold stress in leaves [32]. Interestingly, expression patterns of grape ABA receptors upon abiotic stress were different in leaves and roots. For instance, VvRCAR5 expression was only induced in leaves by salt and cold [32]. However, the functions of most of grape PYL genes in response to abiotic stress remain largely unknown. Additionally, most of previous studies are focused on specific abiotic stress, and the joint influence of different stresses on plant growth is still less studied.
In this study, we explored the expression profiles of grape PYL family in response to cold stress in V. amurensis, and isolated the strongly induced VaPYL4 gene to evaluate its contribution to cold resistance in grape and Arabidopsis. Furthermore, involvement of VaPYL4 in drought and salt tolerance was also demonstrated in Arabidopsis. More importantly, we evaluated the contribution of VaPYL4 to plant growth and development under multiple abiotic stresses (cold, salt and drought) conditions.
Results
VaPYL4 is strongly induced by cold stress
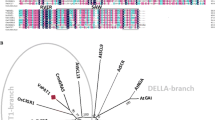
In an attempt to identify cold-responsive PYL genes in grapevine, we analyzed gene expression of the 9 members of PYL family (Fig. 1a) in V. amurensis based on our previous transcriptome data [33]. Among the 9 PYL genes, VaPYL1, VaPYL4, VaPYL5 and VaPYL13 were strongly induced by cold, whereas the expression of VaPYL3 was significantly decreased in response to cold stress (Fig. 1b). Of the four up-regulated genes, VaPYL4 exhibited the highest expression level upon cold stress (Fig. 1b). In addition, we investigated the expression profiles of VaPYL4 in V. amurensis plants in response to cold stress. Two-month-old in vitro V. amurensis plants were treated at 4℃ for 48 h, and grapevine leaves were collected at 0, 2, 4, 8, 12, 24 and 48 h, respectively. The leaves from plants grown at room temperature (25℃) were also sampled at each time point as the controls. The expression of VaPYL4 was detected using quantitative real-time PCR (qPCR). Interestingly, the VaPYL4 gene exhibited changes in expression at 4, 12, 24 and 48 h without cold treatment (Fig. 1c), suggesting that the gene expression might be affected by circadian rhythms. However, the expression of VaPYL4 was significantly increased under cold conditions (from 2 to 48 h) when compared with the corresponding controls (Fig. 1c). Based on these results, we thus selected VaPYL4 as the candidate for further study.
Expression of VaPYL4 is induced by cold stress. A Phylogenetic analysis of PYL ABA receptors from Vitis vinifera and Arabidopsis. The phylogenetic tree was constructed using MEGA5.0 by NJ method with bootstrap replicates of 1000. B Expression profiles of PYLs in V. amurensis in response to cold treatment. The expression data were collected from the previous RNA sequencing results (GSE166247). Data are means of three replicates ± SD. C Expression of VaPYL4 upon cold treatment. Two-month-old in vitro V. amurensis plants were treated at 4℃ for 48 h, and the leaves of cold-treated plants were collected at 0, 2, 4, 8, 12, 24 and 48 h during the cold treatment. The plants grown under normal conditions (25℃) were also sampled as the controls (CK) at each time point. The relative expression level of VaPYL4 was determined by quantitative real-time PCR (qPCR). Data are shown as means ± SD from three biological replicates. Significant differences were determined using Student’s t-test. **P < 0.01
Overexpression of VaPYL4 increases ABA sensitivity
Analysis of PYL4 gene structure revealed that PYL4 contains only one exon, which encodes a protein of 227 amino acids (Additional file 1: Figure S1). The coding sequence (CDS) of PYL4 was amplified from V. amurensis and V. vinifera cv. Pinot Noir, respectively. Alignment of VaPYL4 and VvPYL4 showed that VvPYL4 was totally identical with the reference sequence (‘Pinot Noir’ genome, PN40024), whereas VaPYL4 contained 3 nucleotide changes (Additional file 1: Figure S2). However, analysis of corresponding amino acid sequences uncovered no difference between VaPYL4 and VvPYL4 (Additional file 1: Figure S2). It has been revealed that PYLs function as ABA receptors in nucleus and cytosol [7]. To analyze subcellular localization of VaPYL4, the CDS of VaPYL4 was fused to the enhanced green fluorescent protein (EGFP) gene driven by the cauliflower mosaic virus (CaMV) 35S promoter (Additional file 1: Figure S2). The construct was infiltrated into Nicotiana benthamiana leaves for transient expression through Agrobacterium-mediated transformation. The results showed that the VaPYL4-EGFP fusion protein was localized to nucleus and cytosol (Fig. 2), which is consistent with previous results in Arabidopsis [7].
Subcellular localization of VaPYL4 protein. The coding sequence of VaPYL4 was fused to the N-terminal of EGFP, which was driven by the CaMV 35S promoter. The EGFP fluorescence generated by the 35S::VaPYL4-EGFP construct in epidermal cells of Nicotiana benthamiana leaves was detected by using a confocal laser scanning microscopy. The histone H2B-mCherry was used as an indicator of nucleus. Scale bars correspond to 50 µm
To test the function of VaPYL4 in response to ABA, we cloned the CDS of VaPYL4 into pSAK277 to develop the overexpression vector (Fig. 3a). The VaPYL4-overexpressing (OE) Arabidopsis plants (Additional file 1: Figure S3) were generated by using floral-dip method [34]. The seeds of two transgenic lines (OE5 and OE6) with similar expression level of VaPYL4, as well as the OE9 that exhibited a little higher VaPYL4 expression, were used for germination test on 1/2 Murashige and Skoog (MS) medium supplemented with exogenous ABA. The results showed that there is no significant difference in germination rate between wild-type (WT) and transgenic lines in the absence of exogenous ABA (Additional file 1: Figure S4), which suggested that overexpressing VaPYL4 in Arabidopsis had no much influence on seed germination under normal conditions. However, in the presence of ABA (0.3 μM), the seed germination of transgenic lines was obviously inhibited, exhibiting lower germination rates (40 ~ 44%) when compared with WT (> 87%) (Fig. 3b). Moreover, the inhibitory effect on seed germination was enhanced when ABA concentration was increased (Additional file 1: Figure S4). In addition, the average cotyledon greening rates of OE lines were also much lower than that of WT (Fig. 3c). For instance, the cotyledon greening rate of OE5 in the presence of 0.3 μM ABA was around 37.5%, whereas the greening rate of WT was over 85% (Fig. 3c). All these results suggested that VaPYL4-OE plants are hypersensitive to exogenous ABA.
VaPYL4-overexpressing (OE) plants are hypersensitive to exogenous abscisic acid (ABA). A Schematic illustration of T-DNA region of the pSAK277-VaPYL4 vector for gene overexpression. CaMV35S, cauliflower mosaic virus 35S promoter; TNOS, terminator of nopaline synthase gene; OCS, octopine synthase terminator; PNOS, promoter of nopaline synthase gene; RB, right border; LB, left border. B Seeds germination on 1/2 MS medium containing 0.3 µM ABA. After incubation at 4℃ for 2 d, wild-type (WT) and OE seeds were sown on 1/2 MS medium containing 0.3 µM ABA. The emergence of radicle was measured as germination, and the germination rates were calculated at 3 d after sowing. Scale bars correspond to 0.5 cm. C Cotyledon greening of WT and OE plants on 1/2 MS plates for 6 d. Scale bars correspond to 0.5 cm. Data are collected from three replicates and shown as averages of around 100 seeds. Date are presented as means ± SD. **P < 0.01
Overexpression of VaPYL4 improves cold tolerance of grape callus and Arabidopsis plants
As mentioned above, the expression of VaPYL4 was induced by cold stress (Fig. 1). To investigate the function of VaPYL4 in cold tolerance, we first developed VaPYL4-OE grape calli by introducing the pSAK277-VaPYL4 construct (Fig. 3a) into V. amurensis petioles via Agrobacterium-mediated transformation. After kanamycin-dependent selection, the kanamycin-resistant calli were induced from petiole explants (Fig. 4a). The presence of EGFP reporter gene enabled us to screen transgenic calli rapidly according to the EGFP fluorescence (Fig. 4b). PCR identification was conducted by using NPT II-specific primers (Additional file 1: Table S1), and the results showed that all of the 15 tested grape calli contained exogenous T-DNA insertions (Additional file 1: Figure S5). Furthermore, the results of qPCR revealed that these calli exhibited increased expression levels (> tenfold) of VaPYL4 compared with grape callus transformed with empty vector (EV) (Additional file 1: Figure S5). Three VaPYL4-OE grape calli lines (OE-10, OE-12 and OE-14) with high expression level were selected for subsequent analysis. In addition, we also developed the knockout materials of VaPYL4 by using CRISPR/Cas9 (clustered regulatory interspaced short palindromic repeats/CRISPR-associated protein 9) technology. Two sgRNAs targeting the exon of VaPYL4 were designed and ligated into pCACRISPR/Cas9 vector under the control of VvU6.1 and VvU3.1 promoter, respectively (Fig. 4c). After transformation and antibiotic-dependent selection, two independent calli lines were identified as pyl4 mutants (Fig. 4c). Large fragment deletions (> 50 bp) were detected in the two knockout lines (KO-1 and KO-2), and the mutation efficiencies for KO-1 and KO-2 were 40% and 65%, respectively (Fig. 4c). Low temperature exotherms (LTEs) assay is usually used to evaluate cold tolerance of plant tissues or calli/cells [33, 35, 36]. Thus, the cold tolerance of KO-1 and KO-2, as well as the three OE lines, was evaluated by measuring LTEs using a differential thermal analysis system according to Sun et al. [36]. The LTEs of EV, OE-10 and KO-2, for example, were measured as -5.61, -6.57 and -6.04℃, respectively (Additional file 1: Figure S5). The OE lines exhibited significantly lower LTEs when compared with EV, while the KO lines, however, showed no obvious alterations in LTEs as expected (Fig. 4d). The possible reason is that the presence of wild-type cells in the KO lines might affect the measurement of LTEs. Moreover, multiple PYL genes may function redundantly in cold response, considering that VaPYL1, VaPYL5 and VaPYL13 were also induced by cold treatment (Fig. 1b). These results showed that overexpressing VaPYL4 could enhance cold tolerance of grape calli.
Overexpression of VaPYL4 enhances cold tolerance of grape calli. A Grape calli induced from V. amurensis petioles on selection medium. The V. amurensis petioles were used as explants for co-culture with Agrobacterium cells that contain the VaPYL4 overexpression vector. After co-culture, the V. amurensis petioles were placed on Gamborg medium supplemented with 50 mg/L kanamycin. The induced kanamycin-resistant callus is indicated by red arrow. B Detection of EGFP fluorescence in induced grape calli. C Knockout (KO) of VaPYL4 in grape calli. Two sgRNAs targeting the exon of VaPYL4 were cloned into the pCACRISPR/Cas9 vector. The results of targeted mutagenesis in VaPYL4 in two independent calli (KO-1 and KO-2) were shown. The sequences of sgRNAs are shown in red, while the PAM (protospacer-adjacent motif) sequences are indicated in green. WT, wild-type sequence; Mut, mutated sequence. The number of analyzed amplicons and mutation efficiencies are shown on the right. D Values of low-temperature exotherms (LTEs) of transgenic grape calli. Data are shown as means ± SD collected from at least five biological replicates. **P < 0.01
Freezing treatment was also performed with VaPYL4-OE Arabidopsis plants. Both WT and OE lines were treated at -7℃ for 0.5 h, and the survival rates of OE5, OE6 and OE9 were obviously higher than that of WT (Fig. 5a-b). Moreover, the electrolyte leakage of OE plants was much lower than that of WT after freezing treatment (Fig. 5c). These results showed that ectopic overexpression of VaPYL4 in Arabidopsis could enhance its tolerance to freezing stress. It seems that OE5 and OE6 outperformed OE9 during the freezing treatment (Fig. 5), and the two lines were therefore chosen for subsequent treatments.
Overexpression of VaPYL4 enhances cold tolerance of transgenic Arabidopsis plants. A Freezing phenotypes of WT and OE plants. WT and OE plants were grown on 1/2 MS medium at 22℃ for 10 days before being subjected to freezing treatment. The plates were kept at -7℃ for 0.5 h and then incubated at 4℃ for 12 h. Representative photos were taken after recovery for 3 days under normal conditions. Scale bars correspond to 1 cm. B Survival rates of Arabidopsis plants after freezing treatment. C Electrolyte leakage of Arabidopsis plants after freezing treatment. Data are means ± SD. *P < 0.05; **P < 0.01. Similar results were observed in at least three independent experiments
Overexpression of VaPYL4 enhances the tolerance of Arabidopsis to salt and drought stress
To investigate whether VaPYL4 participates in the response to other abiotic stresses such as salt and drought, we performed salt and drought treatment, respectively, with OE5 and OE6 plants. One-week-old Arabidopsis plants were transferred onto the 1/2 MS medium supplemented with 250 mM mannitol or 150 mM NaCl for treatment. The plant growth of WT and OE lines was obviously inhibited by the presence of mannitol (Fig. 6a), which was used to mimic osmotic or drought stress. Similar phenotypes were also observed for the plants under salt treatment (Fig. 6a). The primary root length of Arabidopsis plants was obviously decreased under stress conditions (Fig. 6b). However, compared with WT, the two transgenic lines, OE5 and OE6, had longer primary roots (Fig. 6b), which suggested that OE5 and OE6 were more resistant to drought and salt stress. Moreover, drought resistance of OE5 and OE6 was further evaluated using pot experiments. The two OE lines were subjected to drought stress and re-watering, and the results showed that the two OE lines were more resistant to drought than WT (Fig. 6c). Compared with an ~ 66.7% survival rate of WT plants, up to 100% of OE5 and OE6 plants survived from a 12-d drought stress treatment followed by a 5-d recovery period. Intriguingly, no WT plants survived after an 18-d drought stress treatment followed by a 5-d recovery period. By contrast, over 69% of OE5 and OE6 plants survived from the treatment (Fig. 6c). All these results showed that overexpression of VaPYL4 enhanced the tolerance of transgenic Arabidopsis to salt and drought stress.
Overexpression of VaPYL4 improves resistance of transgenic Arabidopsis plants to salt and drought stress. A Phenotypes of WT and OE plants under salt (NaCl) and mannitol conditions. Plants growing on normal 1/2 MS medium were shown as the control (ctrl). Scale bars: 1 cm. B Length of primary roots of Arabidopsis plants shown in (A). At least 25 seedlings were used for the measurement for each genotype. C Phenotypes of WT and OE plants under drought conditions. Drought treatment was performed by withholding water for 12 or 15 days, then the plants were re-watered for 5 days. Survival rates were measured after the recovery. Data are means ± SD. **P < 0.01; ***P < 0.001. Similar results were observed in three independent experiments
Overexpression of VaPYL4 reduces adverse effect on plant growth caused by multiple abiotic stresses
In the experiments described above, we evaluated the tolerance of VaPYL4-OE plants to cold, salt and drought stress separately. Nevertheless, as sessile organisms, plants are usually threatened by multiple stresses simultaneously. Based on the results we have obtained, we speculated that overexpressing VaPYL4 may also help to mitigate adverse effect on growth of transgenic plants under conditions of multiple abiotic stresses. To test this hypothesis, 3-week-old seedlings of WT, OE5 and OE6 were first irrigated with 100 mM NaCl solution, and 5 days later the pot seedlings were treated at 4℃ for 3 d, followed by a 14-d drought treatment and 3-d recovery period. The treatment was divided into three different stages (stage 1–3) as shown in Fig. 7a. Stress-induced damages were observed in WT leaves at stage 2 during the treatment (Fig. 7a). Most (about 90%) of WT seedlings were dead at stage 3, while ~ 49% of OE5 and ~ 55.9% of OE6 seedlings successfully survived from the successive treatments of different abiotic stresses (Fig. 7a, c). Moreover, development of the seedlings was also attenuated by the treatment with multiple stresses when compared with those plants grown under normal conditions (Fig. 7b; Additional file 1: Figure S6). However, the growth of OE5 and OE6 seedlings was less affected by multi-stress treatment (Fig. 7b). Though all the seedlings showed a delayed flowering phenotype when treated with abiotic stresses (Fig. 7d), most (33–53%) of transgenic seedlings exhibited an earlier (1–2 d) flowering phenotype when compared with WT plants (Fig. 7d). More importantly, analysis of the fresh weight (FW) of individual seedlings showed that the FW of transgenic plants outweighed the controls (Fig. 7e). In addition, similar results were also observed in the measurement of FW of siliques (Fig. 7f). These results suggested that the OE5 and OE6 plants had more biomass than WT plants after multi-stress treatment.
Overexpression of VaPYL4 improves plant performance after multi-stress treatment. A Phenotypes of WT and OE plants during the treatment with different abiotic stresses. Three-week-old seedlings were first watered with 100 mM NaCl solution for 5 days and then treated at 4℃ for 3 days, followed by a drought treatment by withholding water for 14 days and a 3-day recovery period. Representative images were taken before treatment (stage 1), at 5 days (stage 2) of drought treatment, and at the end of the treatment (stage 3), respectively. The damages observed in WT leaves at stage 2 were indicated by white arrows. Scale bars: 1 cm. B Images showing the development of seedlings and siliques after multi-stress treatment. Photographs were taken at 50 days post-germination. Scale bar: 1 cm. C Survival rates of WT and OE plants. Data are collected from three replicates, and each replicate consists of at least 45 seedlings. D Flowering time of WT and OE plants treated with or without abiotic stresses. The plants with at least one flower were considered to be at the flowering stage. Data are collected from three replicates, and each replicate consists of at least 10 and 50 seedlings for control and multi-stress treatment, respectively. The number of plants survived from treatment and used for flowering time record is shown above the figure. E–F Fresh weight of seedling (E) and siliques (F). The individual plants without roots were used for the measurement of fresh weight (FW). For the measurement of FW of siliques, all the siliques from the same genotype were pooled and a number of 10 siliques were collected as a sample. The replicates from three experiments are shown above the bars. Data are means ± SD. **P < 0.01
Physiological changes were investigated at stage 3 during the treatment. The malondialdehyde (MDA) contents were significantly lower whereas the peroxidase (POD) activities were much higher in OE5 and OE6 plants (Fig. 8a, b). Investigation of expression profiles of stress-responsive genes revealed that the expression levels of RD29A (Responsive to desiccation 29A), COR15A (Cold responsive 15A), COR15B and KIN2 (Kinase 2) in OE5 and OE6 were much higher relative to WT (Fig. 8c). Furthermore, the jasmonic acid (JA) biosynthetic related gene LOX2 (Lipoxygenase 2) and the superoxide gene SOD [37] were also up-regulated in OE5 and OE6 plants (Fig. 8c).
Determination of MDA (malondialdehyde) content, POD (peroxidase) activity and expression of stress-responsive genes. A MDA content measured in WT and OE plants during the multi-stress treatment. B POD activity detected in WT and OE plants during the multi-stress treatment. C Expression of stress-responsive genes in WT and OE plants. The expression levels of stress-responsive genes in OE plants relative to WT were determined by qPCR. Data are mean values ± SD of three biological replicates. *P < 0.05; **P < 0.01
The seedlings at the flowering stage were also used for the experiment. The results showed that Arabidopsis seedlings at the flowering stage were more sensitive to stress treatment, and unsurprisingly, the OE5 and OE6 plants still outperformed the WT plants (Additional file 1: Figure S7). Taken together, our results showed that overexpression of VaPYL4 could help Arabidopsis plants to survive from severe environment conditions and mitigate adverse effect provoked by different abiotic stresses on plant growth.
Discussion
ABA content was generally increased in plants upon abiotic stresses such as drought and salinity [8, 38,39,40], which greatly affect plant growth and distribution [41, 42]. Most of previous studies on ABA-dependent signaling were carried out with a focus on plant response to drought or high salinity [43,44,45,46,47]. Cold tolerance involved in ABA signaling is relatively less studied. Recently, overexpression of PYL genes from Arabidopsis and rice was found to enhance cold tolerance of transgenic plants [28,29,30]. In grapevine, several ABA receptor genes, including VvRCAR5/PYL4 and VvRCAR7, were found to be induced by cold stress [28]. Consistent with this result, the VaPYL4 gene responded to cold treatment as well (Fig. 1). Moreover, the function of VaPYL4 in cold tolerance was further confirmed by gene overexpression in both grape calli and Arabidopsis plants (Fig. 4, 5). These results provide evidence for ABA-mediated cold tolerance in grapevine.
In addition to cold tolerance, drought and salt resistance was also improved in transgenic Arabidopsis plants (Fig. 6), indicating that the VaPYL4 gene has a great potential for broader applications. A recent study reported that overexpression of a wheat ABA receptor increased water-use efficiency and improved grain production under drought condition [48]. The VaPYL4 gene reported here can also serve as a promising candidate for grape and crops improvement. In natural environment, plants need to adapt to different biotic and abiotic stresses. Nevertheless, current studies usually focus on individual abiotic stress. In the present study, we treated the VaPYL4-OE plants with salt, cold and drought stress, and found that the OE5 and OE6 seedlings were less affected by these treatments when compared with WT plants, showing an earlier flowering phenotype, higher survival rate, and heavier weight of seedlings and siliques (Fig. 7). Measurement of MDA content, which is an indicator of plasma membrane damage, showed that OE5 and OE6 plants had much lower level of MDA (Fig. 8a). On the contrary, the activity of POD that helps to scavenge reactive oxygen species generated by abiotic stresses was higher in OE5 and OE6 (Fig. 8b). Regulation of stress-related gene expression is an important mechanism employed by plants to cope with abiotic stresses [2]. Notably, regulatory pathways triggered by different stresses may share the common targets. The COR15A and RD29A gene could be induced by both drought and cold stress [Gene cloning and subcellular localization The leaves of V. amurensis and V. vinifera plants were used to prepare total RNA, which was then adopted for cDNA synthesis by using the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme) following the manufacturer’s instruction. The full-length CDS of PYL4 was amplified from the prepared cDNA libraries of V. amurensis and V. vinifera, respectively, by PCR with the primers PYL4-PCR-F and PYL4-PCR-R (Additional file 1: Table S1) using the KOD-Plus-Neo Kit (TOYOBO). The amplified fragments were cloned into the pLB cloning vector (TIANGEN) for Sanger sequencing assay. To generate the expression vector for subcellular localization, the verified sequence of VaPYL4 without stop codon was amplified from the pLB vector using the primers PYL4-2300-F and PYL4-2300-R and ligated into the modified pCAMBIA2300-EGFP vector through BamHI site via homologous recombination (HR) by using the ClonExpress II One Step Cloning Kit (Vazyme). The VaPYL4 gene driven by the 35S promoter was located at 5’ upstream region of EGFP gene in the 35S::VaPYL4-EGFP vector (Additional file 1: Figure S2). The developed construct was introduced into the Agrobacterium strain GV3101, which was used for infiltration of Nicotiana benthamiana leaves. The fluorescence was detected 3 days after infiltration using Leica TCS SP8 confocal laser scanning microscopy. The histone H2B-mCherry was used as an indicator of nucleus as previously described [33]. To develop the vector for gene overexpression in grape callus and Arabidopsis, the VaPYL4 gene was amplified from the pLB vector using the primers PYL4-P277-F and PYL4-P277-R and ligated into the EcoRI-digested pSAK277-EGFP vector via HR. The overexpression vector was introduced into the Agrobacterium EHA105 and GV3101 for the transformation of grape callus and Arabidopsis, respectively. The transformation of grape callus was conducted by using petioles of V. amurensis plants as the explants according to the protocol described previously [53]. The petioles were placed on Gamborg medium supplemented with 50 mg L−1 kanamycin after co-culture with Agrobacterium cells and sub-cultured monthly until kanamycin-resistant calli were developed. EGFP fluorescence was detected using CCD camera (Tanon 5200) to select transgenic calli rapidly. Then the calli were identified by PCR using NPT II-specific primers (Additional file 1: Table S1). The expression of VaPYL4 in the induced calli was further confirmed by qPCR. Arabidopsis transformation was performed using the floral-dip method [34]. Transgenic plants were screened on 1/2 MS medium with 50 mg L−1 kanamycin. The T3 homozygous transgenic lines were used for the treatments. To knock out VaPYL4 gene in grape, two different targets were designed to target the exon of VaPYL4 using the targetDesign tool of CRISPR-GE (http://skl.scau.edu.cn/targetdesign/). The designed sgRNAs were ligated to grape VvU6.1 and VvU3.1 promoter [54] to develop sgRNA expression cassettes. Then the sgRNA expression cassettes were inserted into pCACRISPR/Cas9 vector [55] through EcoRI and HindIII sites via HR. The construction of the CRISPR vector was carried out as previously described [55]. The well-constructed CRISPR vector was introduced into grape callus by Agrobacterium-mediated transformation [53]. To detect targeted mutagenesis, the induced calli were sampled for genomic DNA extraction, and the DNA fragment containing the target sequence was amplified from genomic DNA. The PCR amplicons were cloned into the pLB vector, and a number of 20 clones were analyzed by Sanger sequencing for each callus. For qPCR assay, the cDNA was synthesized from total RNA using the HiScript II Q RT SuperMix for qPCR Kit (Vazyme). The qPCR was performed using AceQ qPCR SYBR Green Master Mix (Vazyme) with the CFX Manager system (BioRad). The reactions were carried out as described by Ren et al. [56]. Grape Actin 1 and GAPDH [33] and Arabidopsis Actin 2/8 were used as internal controls. Gene expression relative to internal controls was determined using 2−∆∆CT method [57]. Significant differences were determined by Student’s t-test. The plant with at least one flower (Additional file 1: Figure S8) was considered to be at the flowering stage, and the growing time for development of the first flower was recorded as the flowering time for each plant. For the measurement of fresh weight of seedlings, each plant was measured without roots, considering that the soil cannot totally be removed from the roots. To measure the weight of siliques, all the siliques from the same genotype were pooled and a number of 10 siliques were collected as a replicate. The fresh weight was determined at 50 d post-germination. The cold tolerance of grape calli was evaluated by its LTEs, which were measured using the Keithley Multimeter Data Acquisition System (model 2700-DAQ-40) combined with a programmable freezer and a Tenney Environmental Test Chamber (model T2C, Thermal Product Solutions) as previously described [36]. At least 5 biological replicates were used for the measurement. Electrolyte leakage of Arabidopsis plants was analyzed using the method described by Li et al. [57]. Electrolyte leakage assay was repeated three times. To measure the MDA content and POD activity, the plants were ground into powder with liquid nitrogen, and 0.1 g of powder was used for the measurement. The MDA level and POD activity were determined using MDA and POD isolation kits (Solarbio) following the manufacturer’s instructions. Five biological and three technical replicates were conducted.Plant transformation
Targeted mutagenesis of VaPYL4
qPCR assay
Plant phenoty**
Measurement of LTEs and Physiological assays
Availability of data and materials
The transcriptome date used in this study are available in NCBI Gene Expression Omnibus (GEO) database under the accession number of GSE166247. The data generated or used in this study are included in the manuscript and supplementary materials.
Abbreviations
- ABA:
-
Abscisic acid
- CDS:
-
Coding sequence
- COR15A:
-
Cold responsive 15A
- CRISPR/Cas9:
-
Clustered regulatory interspaced short palindromic repeats/CRISPR-associated protein 9
- EGFP:
-
Enhanced green fluorescent protein
- EV:
-
Empty vector
- FW:
-
Fresh weight
- HR:
-
Homologous recombination
- KIN2:
-
Kinase 2
- KO:
-
Knockout
- LTEs:
-
Low temperature exotherms
- MDA:
-
Malondialdehyde
- OE:
-
Overexpression
- POD:
-
Peroxidase
- PYL:
-
Pyrabactin resistance-like proteins
- qPCR:
-
Quantitative real-time PCR
- RD29A:
-
Responsive to desiccation 29A
- WT:
-
Wild type
References
Bansal KC, Lenka SK, Mondal TK. Genomic resources for breeding crops with enhanced abiotic stress tolerance. Plant Breeding. 2014;133:1–11.
Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit Rev Plant Sci. 2005;24:23–58.
Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803.
Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. Pathological hormone imbalances. Curr Opin Plant Biol. 2007;10:372–9.
Lee SC, Luan S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012;35:53–60.
Sah SK, Reddy KR, Li JX. Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci. 2016;7:571.
Dong T, Park Y, Hwang I. Abscisic acid: biosynthesis, inactivation, homoeostasis and signaling. Essays Biochem. 2015;58:29–48.
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–79.
Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Gene Dev. 2010;24:1695–708.
Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y. Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol. 2019;62:25–54.
Brookbank BP, Patel J, Gazzarrini S, Nambara E. Role of basal ABA in plant growth and development. Genes. 2021;12:1936.
Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–80.
Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, et al. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics. 2008;9:550.
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–71.
Saha J, Chatterjee C, Sengupta A, Gupta K, Gupta B. Genome-wide analysis and evolutionary study of sucrose non-fermenting 1-related protein kinase 2 (SnRK2) gene family members in Arabidopsis and Oryza. Comput Biol Chem. 2014;49:59–70.
Huai J, Wang M, He J, Zheng J, Dong Z, Lv H, et al. Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep. 2008;27:1861–8.
Wei K, Pan S. Maize protein phosphatase gene family: identification and molecular characterization. BMC Genomics. 2014;15:773.
Fan W, Zhao M, Li S, Bai X, Li J, Meng H, et al. Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biol. 2016;16:99.
Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q, et al. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J Exp Bot. 2011;62:5659–69.
Boneh U, Biton I, Schwartz A, Ben-Ari G. Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant Sci. 2012;187:89–96.
Zhang R, Wang Y, Li S, Yang L, Liang Z. ABA signaling pathway genes and function during abiotic stress and berry ripening in Vitis vinifera. Gene. 2021;769: 145226.
Gomez-Cadenas A, Vives V, Zandalinas SI, Manzi M, SanchezPerez AM, Perez-Clemente RM, et al. Abscisic acid: a versatile phytohormone in plant signaling and beyond. Cur Protein Pept Sci. 2015;16:413–34.
Ruiz-Partida R, Rosario SM, Lozano-Juste J. An update on crop ABA receptors. Plants. 2021;10:1087.
Zhang H, Zhu J, Gong Z, Zhu JK. Abiotic stress responses in plants. Nat Rev Genet. 2022;23:104–19.
Zhao Y, Chan Z, **ng L, Liu X, Hou YJ, Chinnusamy V, et al. The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res. 2013;23:1380–95.
Shi H, Ye T, Zhu JK, Chan Z. Constitutive production of nitric oxide leads to enhanced drought stress resistance and extensive transcriptional reprogramming in Arabidopsis. J Exp Bot. 2014;65:4119–31.
Zhao Y, Chan Z, Gao J, **ng L, Cao M, Yu C, et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA. 2016;113:1949–54.
Zhang Q, Kong X, Yu Q, Ding Y, Li X, Yang Y. Responses of PYR/PYL/RCAR ABA receptors to contrasting stresses, heat and cold in Arabidopsis. Plant Signal Behav. 2019;14:1670596.
Lenka SK, Muthusamy SK, Chinnusamy V, Bansal KC. Ectopic expression of rice PYL3 enhances cold and drought tolerance in Arabidopsis thaliana. Mol Biotechnol. 2018;60:350–61.
Verma RK, Santosh-Kumar VV, Yadav SK, Pushkar S, Rao MV, Chinnusamy V. Overexpression of ABA receptor PYL10 gene confers drought and cold tolerance to indica rice. Front Plant Sci. 2019;10:1488.
Li G, **n H, Zheng XF, Li S, Hu Z. Identification of the abscisic acid receptor VvPYL1 in Vitis vinifera. Plant Biol. 2012;14:244–8.
Boneh U, Biton I, Zheng C, Schwartz A, Ben-Ari G. Characterization of potential ABA receptors in Vitis vinifera. Plant Cell Rep. 2012;31:311–21.
Ren C, Li H, Wang Z, Dai Z, Lecourieux F, Kuang Y, et al. Characterization of chromatin accessibility and gene expression upon cold stress reveals the transcription factor RAV1 functions in cold response in Vitis amurensis. Plant Cell Physiol. 2021;62:1615–29.
Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006;1:641–6.
Mills LJ, Ferguson JC, Keller M. Cold-hardiness evaluation of grapevine buds and cane tissues. Am J Enol Viticult. 2006;57:194–200.
Sun X, Matus JT, Wong DCJ, Wang Z, Chai F, Zhang L, et al. The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promoters the accumulation of raffinose family oligosaccharides. J Exp Bot. 2018;69:1749–64.
Fang L, Su L, Sun X, Li X, Sun M, Karungo SK, et al. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J Exp Bot. 2016;67:2829–45.
Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003;133:1755–67.
Christmann A, Weiler EW, Steudle E, Grill E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 2007;52:167–74.
Endo A, Sawada Y, Takahashi H, Okamoto M, Ikegami K, Koiwai H, et al. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 2008;147:1984–93.
Parihar P, Singh S, Singh R, Singh VP, Prasad SM. Effect of salinity stress on plants and its tolerance strategies: a review. Environ Sci Pollut Res Int. 2015;22:4056–75.
Ullah A, Sun H, Yang XY, Zhang XL. Drought co** strategies in cotton: increased crop per drop. Plant Biotechnol J. 2017;15:271–84.
Kim S, Kang JY, Cho DI, Park JH, Kim SY. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004;40:75–87.
Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470–88.
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–85.
Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124:509–25.
Luo X, Li C, He X, Zhang X, Zhu L. ABA signaling is negatively regulated by GbWRKY1 through JAZ1 and ABI1 to affect salt and drought tolerance. Plant Cell Rep. 2020;39:181–94.
Mega R, Abe F, Kim JS, Tsuboi Y, Tanaka K, Kobayashi H, et al. Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat Plants. 2019;5:153–9.
**ong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–83.
Liu L, Gallagher J, Arevalo ED, Chen R, Skopelitis T, Wu Q, et al. Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat Plants. 2021;7:287–94.
Song X, Meng X, Guo H, Cheng Q, **g Y, Chen M, et al. Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat Biotechnol. 2022. https://doi.org/10.1038/s41587-022-01281-7.
Zhang D, Hussain A, Manghwar H, **e K, **e S, Zhao S, et al. Genome editing with the CRISPR-Cas system: an art, ethics and global regulatory perspective. Plant Biotechnol J. 2020;18:1651–69.
Zhao T, Wang Z, Su L, Sun X, Cheng J, Zhang L, et al. An efficient method for transgenic callus induction from Vitis amurensis petiole. PLoS ONE. 2017;12: e0179730.
Ren C, Liu Y, Guo Y, Duan W, Fan P, Li S, et al. Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Hortic Res. 2021;8:52.
Ren C, Liu X, Zhang Z, Wang Y, Duan W, Li S, et al. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L). Sci Rep. 2016;6:32289.
Ren C, Zhang Z, Wang Y, Li S, Liang Z. Genome-wide identification and characterization of the NF-Y gene family in grape (Vitis vinifera L.). BMC Genomics. 2016;17:605.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods. 2001;25:402–8.
Li H, Ding Y, Shi Y, Zhang X, Zhang S, Gong Z, et al. MPK3-and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev Cell. 2017;43:630–42.
Acknowledgements
This study was conducted as part of the Franco-Sino LIA INNOGRAPE International Associated Laboratory.
Funding
This work was jointly funded by grants from the National Natural Science Foundation of China (32001994 and 32025032), National Key Research and Development Program of China (2019YFD1002501), and Agricultural Breeding Project of Ningxia Hui Autonomous Region (NXNYYZ20210104).
Author information
Authors and Affiliations
Contributions
CR and ZL conceived and designed the experiments; CR, YK, YL and YG performed the experiments, CR and HL performed data analysis; CR wrote the manuscript; PF, SL and ZL revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ren, C., Kuang, Y., Lin, Y. et al. Overexpression of grape ABA receptor gene VaPYL4 enhances tolerance to multiple abiotic stresses in Arabidopsis. BMC Plant Biol 22, 271 (2022). https://doi.org/10.1186/s12870-022-03663-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-022-03663-0