Abstract
Background
Resistance to pre-harvest sprouting (PHS) is one of the major objectives in wheat breeding programs. However, the complex quantitative nature of this trait presents challenges when breeding for PHS resistance. Characterization of PHS using near-isogenic lines (NILs) targeting major quantitative trait locus/loci (QTL/QTLs) can be an effective strategy for the identification of responsible genes and underlying mechanisms.
Results
In this study, multiple pairs of NILs were developed and confirmed for a major QTL located on the 4BL chromosome arm that contributes to PHS resistance in wheat, using a combined heterogeneous inbred family method and a fast generation cycling system. Phenotypic characterization of these confirmed NILs revealed significant differences in PHS resistance between the isolines, where the presence of the resistant allele increased the resistance to sprouting on spikes by 54.0–81.9% (average 70.8%) and reduced seed germination by 59.4–70.5% (average 66.2%). The 90 K SNP genoty** assay on the confirmed NIL pairs identified eight SNPs on 4BL, associated with five candidate genes; two of the candidate genes were related to seed dormancy. Agronomic traits in the NIL pairs were investigated; both plant height and grain number per spike were positively correlated with PHS susceptibility.
Conclusions
This study confirmed multiple pairs of NILs and identified SNPs between PHS isolines, which are valuable resources for further fine-map** of this locus to clone the major genes for PHS resistance and investigate the possible functional regulation of these genes on important agronomic traits, such as plant height and grain number per spike.
Similar content being viewed by others
Background
Pre-harvest sprouting (PHS) is a phenomenon in which seeds germinate on the intact spike or on the mother plant before harvest. PHS is triggered when matured grains encounter rain or air humidity close to saturation. In wheat, PHS results in degradation of starches in the grain rendering the grain unsuitable for milling into flour [1]. PHS occurs in many countries including China, Australia, Canada, USA, Europe and Japan [7,8]. PHS is due to insufficient grain dormancy during seed development and maturation [9] and its resistance can be affected by factors including substances in the glume that inhibit germination [10], physical barriers to water penetration in the spike [11], and spike morphology [12]. Therefore, germination testing directly on a spike (spike sprouting test) reflects PHS performance better than a standard seed germination test [13].
PHS resistance is a quantitative trait that is regulated by several genes or quantitative trait loci (QTLs) [14]. QTLs for PHS resistance have been reported on almost all chromosomes in bread wheat [14,15,16]; several major QTLs have been located on group 4 chromosomes with repeated detections in different studies [17,18,19,20,21,22,23,24,25,26,27]. The genes underlying the major PHS resistance QTL on 4AL have been recently identified [8]. Despite the considerable attention paid to the 4AL QTL, little research has been conducted to characterize other major QTLs on group 4 chromosomes. A QTL within the centromeric region of the long arm of chromosome 4B, QPhs.ocs-4B.1, located between markers Xgwm495 and Xgwm375, was identified as a major QTL for grain dormancy in the medium-dormant wheat Chinese Spring, explaining about 20% of the phenotypic variation [19]. Kato et al. [17] identified three QTLs on group 4 chromosomes in doubled-haploid lines, of which Qphs.ocs-4B.2, located within the telomere region of 4BL, had consistent effects on seed dormancy in seeds produced in both glasshouse and growth chamber trials, and was homoeologous with a PHS QTL on 4D. Kumar et al. [25] located a major QTL, QPhs.spa-4B, explaining 35–60% of the phenotypic variation in field environments. Furthermore, using composite interval map**, Lin et al. [26] detected a minor QTL on 4B, Qphs.pseru-4B.1, between markers Xbarc20 and Xwmc238, for both PHS resistance and seed dormancy, which explained 6.3–8.7% of the phenotypic variance.
In the past two decades, QTLs on chromosome 4B have gained attention, as numerous studies have identified a ‘QTL-hotspot’ region on the chromosome responsible for many agronomic traits, especially plant height and yield-related traits [30,31,33,34]. For plant height, the Rht-B1 gene on chromosome 4B has a profound impact on stem elongation and grain number per spike [29]. Marza et al. [31] consistently detected QTLs that influence plant height, spike length and grain number per spike on chromosome 4B in their nine experimental environments. Wang et al. [35] identified a major QTL for grain number per spike on chromosome 4B, and its contribution explained 18.2% of the phenotypic variation. Recently, Guan et al. [36]. NILs are a pair of plants with a nearly identical genetic background except for a single section/locus on one chromosome. This means that any differences between the pairs will be due to differences in genes at the target locus, turning a complex quantitative trait into a simple Mendelian factor with predictable patterns of segregation and inheritance. Generating more than one NIL pair from different initial crosses ensures that the target gene/locus and its function are validated under different genetic backgrounds. Here, we report the development, confirmation, and genotypic and phenotypic characterization of five pairs of NILs targeting the major 4BL QTL, QPhs.ocs-4B.1, responsible for PHS resistance in wheat. Correlations between PHS and plant height, and grain number per spike were also investigated.
Results
Development of NILs for PHS resistance
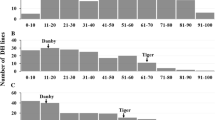
Molecular marker screening of the two cross-population lines with Xgwm495 grouped the progeny into three distinct classes: homozygous with PHS resistant allele of 175 bp, homozygous with susceptible allele of 190 bp, and heterozygous with both 175 bp and 190 bp alleles. Heterozygous lines were selected from each generation for further advancement and marker screening until F7. The marker-assisted heterogeneous inbred family (HIF) method in the two populations generated 19 F7 heterozygous lines—seven from the SUN326AE × Westonia population and 12 from the Chara × DM5637B*8 population. From each heterozygous F7 line, two homozygous F8 isolines with contrasting alleles at marker Xgwm495 were selected as a NIL pair. The 19 pairs of NILs were ranked for their PHS resistance following the sprouting test assay. Selected NIL pairs and their allele categories are shown in Fig. 1.
Agarose gel image of PCR products amplified from the parents and the confirmed isolines using maker Xgwm495. The 175 bp-fragments represent the resistant allele, while the 190 bp fragments represent the susceptible allele. M: 100 bp DNA ladder; numbers 1 to 20 refer to the four parents (Chara, Westonia, DM5637B*8 and SUN326AE) and the eight confirmed isoline pairs as indicated in Table 1
Evaluation of NILs using sprouting test
The sprouting test, using whole spikes, was conducted to analyze PHS resistance in the 19 putative NIL pairs. Differences in sprouting were observed between the resistant and susceptible isolines from day 3 of the test. For the susceptible spikes, young leaves and roots started to grow out of the wrapped filter paper. After removing the wrapped filter paper on day 7, numerous seedlings were observed on each spike of the susceptible isolines, but none or few were observed on each spike of the resistant isolines (Fig. 2). However, drying and threshing of the resistant isoline spikes detected some additional germinated seeds, as their seed coats were broken, and embryos started to grow. Statistical analysis showed significant differences in sprouting in eight of the 19 putative NIL isoline pairs (Table 1).
Two of the eight confirmed NIL pairs—NIL_PHSR4BL_1 and NIL_PHSR4BL_2—were derived from the SUN326AE × Westonia population, and the remaining six—NIL_PHSR4BL_3, NIL_PHSR4BL_4, NIL_PHSR4BL_5, NIL_PHSR4BL_6, NIL_PHSR4BL_7, and NIL_PHSR4BL_8 were derived from the Chara × DM5637B*8 population.
In general, isolines possessing the allele from the resistant parents had lower sprouting percentages than their respective counterparts for all eight NIL pairs. Sprouting percentages of the NILs from SUN326AE × Westonia population ranged from 20 to 73%. Isolines with the resistant allele (NIL_PHSR4BL_1R and NIL_PHSR4BL_2R) had an average sprouting percentage of 27.5%, which was significantly lower than the isoline with the susceptible allele (73.3%). Similarly, the six NIL pairs derived from the Chara × DM5637B*8 population varied considerably in terms of sprouting percentage. Isolines with the susceptible allele had sprouting percentages ranging from 39.2–58.9% (average 54.2%), more than threefold higher than isolines with the resistant allele, (7.1–21.2%). Of the confirmed NIL pairs, NIL_PHSR4BL_2 had the least significant difference between isolines for sprouting percentage (54.0%), while NIL_PHSR4BL_7 had the greatest difference (81.9%).
Evaluation of NILs using GI assay
Germination index (GI) assay was conducted to analyze seed dormancy in the eight putative NIL pairs that had significant differences between the resistant and susceptible isolines in the sprouting test. Assay observation showed that seeds from all the isolines started germination on the second day of the test. For the isolines with susceptible alleles, more than half of the seeds germinated within 2 days, and all seeds had germinated before day 5. In contrast, for the resistant alleles, most of the seed germination occurred evenly from day 2 to day 4; some of the seeds even germinated on the day 6 or 7. GI analysis indicated that five of the eight pairs showed significant differences in GI between the isolines (Table 1). Two (NIL_PHSR4BL_1 and NIL_PHSR4BL_2) of these five NIL pairs were derived from the ‘SUN326AE × Westonia’ population, and the remaining three pairs (NIL_PHSR4BL_4, NIL_PHSR4BL_5, and NIL_PHSR4BL_7) were from ‘Chara × DM5637B*8’ population. Without exception, isolines with the allele from the resistant parent, ‘SUN326AE’ or ‘DM5637B*8’, always had a significantly lower GI than their counterparts for all of the five pairs of NILs. The increase of seed dormancy received from the resistant allele from ‘SUN326AE’ ranged from 59.4–61.6% (average 60.5%) and from ‘DM5637B*8’ ranged from 69.5–70.5% (average 70.1%).
Of the eight NIL pairs with significant differences between resistant and susceptible isolines in the sprouting test, three pairs (NIL_PHSR4BL_3, NIL_PHSR4BL_6 and NIL_PHSR4BL_8) had similarly high GIs between the isolines (0.67–0.77, average 0.71).
Correlations of PHS with other agronomic traits
Of the 11 agronomic traits investigated, only two were significantly correlated with PHS. Figure 3 shows positive correlation in the eight confirmed NILs between sprouting percentage and plant height (r2 = 0.34, p < 0.05), and grain number per spike (r2 = 0.73, p < 0.001), but no significant correlations between sprouting percentage and the other nine traits.
Further investigation showed significant differences between some of the NIL pairs for plant height and grain number per spike (Fig. 4). The resistant and susceptible isolines of four pairs of NILs, namely, NIL_PHSR4BL_1, NIL_PHSR4BL_2, NIL_PHSR4BL_5 and NIL_PHSR4BL_7, differed significantly (p < 0.01) for plant height (Fig. 2): the isolines with resistant allele were shorter than those with susceptible alleles. The NILs with resistant alleles, namely, NIL_PHSR4BL_1, NIL_PHSR4BL_3, NIL_PHSR4BL_5, and NIL_PHSR4BL_6, had significantly fewer grains per spike than those with susceptible alleles. Although the remaining NILs showed no significant differences between the resistant and susceptible isolines for plant height or grain number per spike, isolines with the susceptible allele were generally taller or produced more grains per spike than those with the resistant allele.
SNP genoty** analysis
Five confirmed NIL pairs, with significant differences between resistant and susceptible isolines in both the sprouting test and GI assay, were subjected to 90 K iSelect genoty** assay. The 90 K iSelect genoty** assay was developed based on hexaploid wheat and Aegilops tauschii sequences [37]. Of the 81,587 SNPs on the array, 31,807 were analyzed across the 21 chromosomes after removing those that did not meet selection criteria and those without polymorphism. A total of 989 SNPs were assigned on the long arm of chromosome 4B according to the assembly results from Wang et al. [37]. Analyses of the 989 SNPs among the five confirmed pairs of isolines found that only eight SNPs, which were all located within the 4BL QTL region, showed consistent contrasting genotypes (AA or BB) between the resistant or susceptible isolines. Of the eight SNPs, BS00068851_51 was located between two genes TraesCS4B01G434700LC and TraesCS4B01G434800LC at their untranslated regions (UTRs); BS00096604_51 and IAAV5117, Ra_c27465_569 and Ra_c27465_564 were blasted on the same candidate genes TraesCS4B01G239700 and TraesCS4B02G253300, respectively. Therefore five candidate genes were positioned within the region of QPhs.ocs-4B.1 by blasting the reference Chinese Spring genome (https://triticeaetoolbox.org/wheat//) (Table 2). Functional annotation of these genes indicated that TraesCS4B01G239700 was a heat shock transcription factor and TraesCS4B02G247900 was a DEAD-box ATP-dependent RNA helicase gene, which have been reported to be related to PHS resistance [38,39,40,41,7,8, 14]; while seed dormancy is a major genetic factor of PHS resistance, other factors could be involved in the overall PHS resistance such as glume substances that inhibit germination [10], physical barriers to water penetration in the spike [11] and spike morphology [12]. The three NIL pairs showing significant differences in the sprouting test but similar high GI rates in this study could be valuable materials for understanding how those factors influence PHS resistance.
Apart from PHS phenoty**, we also measured other agronomic traits to investigate possible relationships between PHS and these traits. Statistical analysis of the NIL population showed positive correlation between PHS susceptibility and plant height and grain number per spike, two complex quantitative traits in wheat that are strongly influenced by interacting genetic and environmental factors. Numerous studies in the past two decades have identified QTLs related to plant height or yield-related components on all 21 wheat chromosomes [30, 35]. Notably, for both plant height and grain number per spike, consistent QTLs across multiple genetic backgrounds have been identified on chromosome 4BL. The dwarfing gene Rht-B1, which can increase grain yield and reduce plant height [29], is located on the short arm of chromosome 4B [43]. The QTL QPhs.ocs-4B.1 targeted in this study is located on the long arm of chromosome 4B [19], suggesting the possibility of another gene(s) regulating plant height and grain number per spike, as well as PHS resistance.
Some studies have reported negative correlation between plant height and grain number per spike or yield, especially in plants with the dwarfing gene Rht1 [49], assisted by a fast generation cycling system [50, 51]. The HIF method is useful for develo** multiple sets of NILs with different genetic backgrounds from a single cross, while the fast generation cycling system dramatically shortens the life-cycle by using young embryo culture and promoting flower differentiation. Combining HIF with the fast generation system and marker-assisted selection enabled the fast development of NILs with multiple genetic backgrounds. This approach has been successful in the development of NILs for some major QTL in wheat [52,53,54] and barley [55].
This research adopted SNP assays using the wheat 90 K Illumina iSelect chip to detect potential genes responsible for PHS resistance. Eight SNPs with consistent contrasting genotypes across the five confirmed NIL pairs were identified and associated with five candidate genes located on chromosome 4B. Of the five functional genes, TraesCS4B01G239700 was described as a heat shock transcription factor (Hsf). A study in Arabidopsis thaliana indicated that a specialized Hsf, HsfA9, exclusively expressed in later stages of seed development, coincided with the acquisition of dormancy, and response to abscisic acid [38]. Almoguera et al. [39] detected HsfA9 expressed in embryos, but it disappeared shortly after seed germination in sunflower. In addition, HsfA9 had minor effects on seed maturation and germination [53]. Another candidate gene TraesCS4B02G247900 was described as an ATP-dependent RNA helicase. Recently, a study identified a DEAD-box helicase ATP-binding protein from rice in the response to ABA stress, which is the main plant hormone that regulates germination [41]. A study on DEAD-box RNA helicase showed its influence on germination at high temperature in Arabidopsis seeds [ NIL pairs were developed from two biparental populations including 200 F2 plants of Chara × DM5637B*8 and 40 F2 plants of SUN326AE × Westonia. DM5637B*8 and SUN326AE are the two PHS resistance genotypes, whereas Westonia and Chara are the two Australian non-dormant, PHS susceptible varieties [18, 22, 59, 60]. Seeds of the four parents were obtained from Australian Grains Genebank, Horsham, Victoria, Australia. Molecular marker-assisted HIF method [49], in combination with a fast generation cycling system [50] was used to develop NILs from these two segregating populations. Heterozygous progenies identified from marker-assisted selection (as described in the below “Molecular marker analysis” section) were selfed for the next generation. In detail, 240 F2 lines were planted in the glasshouse. Around 10 days after anthesis, young embryos from each plant were germinated on an artificial medium for next generation [50]. At least four young seedlings from each plant were transferred into pot for further growing. Only heterozygous progenies, identified using marker-assisted selection at three-leaf stage, were kept to grow into next generation for the next round of selection. This heterozygous progeny selection and selfing progress was continued until the F7 generation. For F8 individuals, either homozygous with the resistant allele from SUN326AE or DM5637B*8, or with the susceptible allele from Westonia or Chara, were identified from the progenies of F7 heterozygous plants as putative NIL pairs. Seeds from these putative NIL pairs were multiplied, and their PHS responses were assessed to identify the true NILs. The two populations were grown in controlled environments in tissue culture rooms and glasshouses at The University of Western Australian in Perth, with DNA marker-assisted selection in each generation. Genomic DNA was isolated from two-week-old seedlings using a modified CTAB method [61] for each plant in every generation. SSR marker Xgwm495 (Forward: GAGAGCCTCGCGAAATATAGG; Reverse: TGCTTCTGGTGTTCCTTCG), located close to QPhs.ocs-4B.1, was used to identify heterozygous progeny from the two populations. PCR reactions were performed and amplification products were viewed following the protocol described in Wang et al. [35]. At physiological maturity, as the head and the peduncle lost their green color (about 30 days after flowering), four spikes from each plant were harvested for PHS assessment. Three plants as three replicates for each putative isoline were tested. The spikes were wrapped in moist filter paper immediately after being immersed in distilled water for 6 h, and then randomly arranged standing upright in a sealed box with 100% humidity for 7 days. The box was placed in a controlled temperature room with a constant condition of 22 °C and 16 h light/8 h dark. After 7 days, the spikes were immediately dried at 65 °C to prevent further germination and then gently threshed to collect grains, which were examined for symptoms of sprouting damage (breakage of the seed coat near the embryo). The sprouting percentage was calculated as the percentage of sprouted grains to the total number of grains, as described in Shorinola et al. [62]. GI assay was conducted on all putative pairs of the isolines. Seeds for the GI assay were harvested at the same time as the spikes for the sprouting test, and gently threshed to keep uniform seed condition. Three replicates were used for each genotype. For each replicate, 50 healthy seeds were evenly distributed in a 90 mm Petri dish with deionized water and filter paper for 7 days. The Petri dish were sealed with Parafilm to prevent water evaporation. Germinated seeds were counted daily and then removed. The germination index was calculated as follows [63]: where n1, n2,…, n7 are the number of seeds germinated on the first, second, and subsequent days until the 7th day, respectively. The maximum index is 1.0 if all grains germinate on the first day. To investigate possible correlations of other agronomic traits with PHS resistance, measurements of various traits were taken during plant growth, including days to flowering, days to physiological maturity, plant height, plant dry matter, spike number, grain per spike, yield per plant, and 1000-kernel weight. Days to flowering for each plant was recorded as the date when half of the spikes were flowered; days to physiological maturity was recorded as the head and the peduncle lost their green color. Plant height was measured from the ground to the tip of the tallest tiller of the plant. Plant dry matter was determined after the plants were harvested and oven dried at 80 °C to constant weight; and then spike number, grain per spike, yield per plant and 1000-kernel weight were measured for each plant. Statistical analysis was performed using GenStat statistical software 17th edition. The sprouting percentages from each putative NIL pairs were analysed using the Student’s t-test to determine any significant differences between the isoline pairs with or without the resistant allele from the resistant genotypes SUN326AE and DM5637B*8. Correlation analysis was conducted to investigate possible morphological traits correlated with PHS resistance. Genomic DNA samples of the confirmed NILs were genotyped by the custom Illumina wheat 90 K iSelect Assay [37]. SNP clustering and genotype calling were performed using GenomeStudio 2.0 software (Illumina). SNPs with a call frequency < 0.8 (i.e. missing data points > 20%) or minor allele frequency (MAF) < 0.05 and SNPs with > 0.25 heterozygous calls were removed. The sequences of the SNPs that differed between the NIL pairs were used to perform a BLAST search against the wheat reference genome [64]. The SNPs located on the 4BL chromosome arm, especially those near markers Xgwm495 and Xgwm375, were scrutinized for candidate genes underlying the targeted QTL.Methods
Plant materials
Molecular marker analysis
Sprouting test
Germination index assay
Investigation of the correlation between PHS resistance and other agronomic traits
Statistical analysis
90 K SNP genoty** and candidate gene identification
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- GI:
-
Germination index
- HIF:
-
Heterogeneous inbred family
- Hsf:
-
Heat shock transcription factor
- NIL:
-
Near-isogenic line
- PHS:
-
Pre-harvest sprouting
- QTL:
-
Quantitative trait locus/loci
- SNP:
-
Single nucleotide polymorphism
References
Wahl TI, Desmond O'Rourke A. The economics of sprout damage in wheat. Agribusiness. 1994;10(1):27–41.
Yang Y, Zhao X, **a L, Chen X, **a X, Yu Z, He Z, Roder M. Development and validation of a Viviparous-1 STS marker for pre-harvest sprouting tolerance in Chinese wheats. Theor Appl Genet. 2007;115(7):971–80.
Biddulph T, Plummer J, Setter T, Mares D. Seasonal conditions influence dormancy and preharvest sprouting tolerance of wheat (Triticum aestivum L.) in the field. Field Crop Res. 2008;107(2):116–28.
Biddulph T. Mechanisms of dormancy, preharvest sprouting tolerance and how they are influenced by the environment during grain filling and maturation in wheat (Triticum aestivum L.). Perth: The University of Western Australia; 2006. https://research-repository.uwa.edu.au/en/publications/mechanisms-of-dormancy-preharvest-sprouting-tolerance-and-how-the. PhD thesis
Darabi H, Mohandessi S, Balavar Y, Moghaddam M, Aghapoor K, Mohsenzadeh F, Nourinia A. Clove bud oil: an efficient, economical and widely available oil for the inhibition of wheat seed germination. Environ Chem Lett. 2011;9(4):519–24.
Derera N, Bhatt G, McMaster G. On the problem of pre-harvest sprouting of wheat. Euphytica. 1977;26:299–308.
Torada A, Koike M, Ikeguchi S, Tsutsui I. Map** of a major locus controlling seed dormancy using backcrossed progenies in wheat (Triticum aestivum L.). Genome. 2008;51(6):426–32.
Barrero JM, Cavanagh C, Verbyla KL, Tibbits JF, Verbyla AP, Huang BE, Rosewarne GM, Stephen S, Wang P, Whan A, et al. Transcriptomic analysis of wheat near-isogenic lines identifies PM19-A1 and A2 as candidates for a major dormancy QTL. Genome Biol. 2015;16:93.
Tuttle KM, Martinez SA, Schramm EC, Takebayashi Y, Seo M, Steber CM. Grain dormancy loss is associated with changes in ABA and GA sensitivity and hormone accumulation in bread wheat, Triticum aestivum (L.). Seed Sci Res. 2015;25(2):179–93.
Salmon D, Helm J, Duggan T, Lakeman D. The influence of chaff extracts on the germination of spring triticale. Agron J. 1986;78(5):863–7.
Gale M. The genetics of preharvest sprouting in cereals, particularly in wheat. In: Derera NF, editor. Preharvest field sprouting in cereals. Boca Raton: CRC Press; 1989.
King R. Water uptake in relation to pre-harvest sprouting damage in wheat: grain characteristics. Aust J Agric Res. 1984;35(3):337–45.
Paterson AH, Sorrells ME, Obendorf RL. Methods of evaluation for preharvest sprouting resistance in wheat breeding programs. Can J Plant Sci. 1989;69(3):681–9.
Anderson J, Sorrells M, Tanksley S. RFLP analysis of genomic regions associated with resistance to pre-harvest sprouting in wheat. Crop Sci. 1993;33:453–9.
Mares D, Mrva K. Map** quantitative trait loci associated with variation in dormancy in Australian wheat. Aust J Agric Res. 2001;52:1257–65.
Mares D, Mrva K, Tan M, Sharp P. Dormancy in white-grained wheat progress towards identification of genes and molecular markers. Euphytica. 2002;126:47–53.
Kato K, Nakamura W, Tabiki T, Miura H, Sawada S. Detection of loci controlling seed dormancy on group 4 chromosomes of wheat and comparative map** with rice and barley genomes. Theor Appl Genet. 2001;102(6):980–5.
Lohwasser U, Röder MS, Börner A. QTL map** of the domestication traits pre-harvest sprouting and dormancy in wheat (Triticum aestivum L.). Euphytica. 2005;143(3):247–9.
Mori M, Uchino N, Chono M, Kato K, Miura H. Map** QTLs for grain dormancy on wheat chromosome 3A and the group 4 chromosomes, and their combined effect. Theor Appl Genet. 2005;110(7):1315–23.
Chen C, Cai S, Bai G. A major QTL controlling seed dormancy and pre-harvest sprouting resistance on chromosome 4A in a Chinese wheat landrace. Mol Breed. 2008;21(3):351–8.
Imtiaz M, Ogbonnaya F, Oman J, Ginkel M. Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines. Genetics. 2008;178(3):1725–36.
Zhang X-Q, Li C, Tay A, Lance R, Mares D, Cheong J, Cakir M, Ma J, Appels R. A new PCR-based marker on chromosome 4AL for resistance to pre-harvest sprouting in wheat (Triticum aestivum L.). Mol Breed. 2008;22(2):227–36.
Cabral A, Jordan M, McCartney C, You F, Humphreys D, MacLachlan R, Pozniak C. Identification of candidate genes, regions and markers for pre-harvest sprouting resistance in wheat. BMC Plant Biol. 2014;14:340–51.
Albrecht T, Oberforster M, Kempf H, Ramgraber L, Schacht J, Kazman E, Zechner E, Neumayer A, Hartl L, Mohler V. Genome-wide association map** of preharvest sprouting resistance in a diversity panel of European winter wheats. J Appl Genet. 2015;56(3):277–85.
Kumar S, Knox R, Clarke F, Pozniak C, DePauw R, Cuthbert R, Fox S. Maximizing the identification of QTL for pre-harvest sprouting resistance using seed dormancy measures in a white-grained hexaploid wheat population. Euphytica. 2015;205(1):287–309.
Lin M, Cai S, Wang S, Liu S, Zhang G, Bai G. Genoty**-by-sequencing (GBS) identified SNP tightly linked to QTL for pre-harvest sprouting resistance. Theor Appl Genet. 2015;128(7):1385–95.
Torada A, Koike M, Ogawa T, Takenouchi Y, Tadamura K, Wu J, Matsumoto T, Kawaura K, Ogihara Y. A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr Biol. 2016;26(6):782–7.
Zhai H, Feng Z, Li J, Liu X, **ao S, Ni Z, Sun Q. QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front Plant Sci. 2016;7:1617.
Youssefian S, Kirby EJM, Gale MD. Pleiotropic effects of the GA-insensitive Rht dwarfing genes in wheat. 2. Effects on leaf, stem, ear and floret growth. Field Crop Res. 1992;28(3):191–210.
Patil RM, Tamhankar SA, Oak MD, Raut AL, Honrao BK, Rao VS, Misra SC. Map** of QTL for agronomic traits and kernel characters in durum wheat (Triticum durum Desf.). Euphytica. 2013;190(1):117–29.
Marza F, Bai GH, Carver BF, Zhou WC. Quantitative trait loci for yield and related traits in the wheat population Ning7840× Clark. Theor Appl Genet. 2006;112(4):688–98.
Guan P, Lu L, Jia L, Kabir MR, Zhang J, Zhao Y, **n M, Hu Z, Yao Y, Ni Z. Global QTL analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). Front Plant Sci. 2018;9:529.
Zhang H, Chen J, Li R, Deng Z, Zhang K, Liu B, Tian J. Conditional QTL map** of three yield components in common wheat (Triticum aestivum L.). Crop J. 2016;4(3):220–8.
Dan C, Wu X-Y, Kuo WU, Zhang J-P, Liu W-H, Yang X-M, Li X-Q, Lu Y-Q, Li L-H. Novel and favorable genomic regions for spike related traits in a wheat germplasm Pubing 3504 with high grain number per spike under varying environments. J Integr Agric. 2017;16(11):2386–401.
Wang J, Liu W, Wang H, Li L, Wu J, Yang X, Li X, Gao A. QTL map** of yield-related traits in the wheat germplasm 3228. Euphytica. 2011;177(2):277–92.
Somyong S, Ishikawa G, Munkvold J, Tanaka J, Benscher D, Cho Y, Sorrells M. Fine map** of a preharvest sprouting QTL interval on chromosome 2B in white wheat. Theor Appl Genet. 2014;127(8):1843–55.
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J. 2014;12(6):787–96.
Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell. 2007;19(1):182–95.
Almoguera C, Rojas A, Dıaz-Martın J, Prieto-Dapena P, Carranco R, Jordano J. A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. J Biol Chem. 2002;277(46):43866–72.
Tejedor-Cano J, Prieto-Dapena P, Almoguera C, Carranco R, Hiratsu K, Ohme-Takagi M, Jordano J. Loss of function of the HSFA9 seed longevity program. Plant Cell Environ. 2010;33(8):1408–17.
Macovei A, Vaid N, Tula S, Tuteja N. A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signal Behav. 2012;7(9):1138–43.
Gong Z, Dong C-H, Lee H, Zhu J, **ong L, Gong D, Stevenson B, Zhu J-K. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell. 2005;17(1):256–67.
Sourdille P, Charmet G, Trottet M, Tixier MH, Boeuf C, Negre S, Barloy D, Bernard M. Linkage between RFLP molecular markers and the dwarfing genes Rht-B1 and Rht-D1 in wheat. Hereditas. 1998;128(1):41–6.
Flintham JE, Börner A, Worland AJ, Gale MD. Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J Agric Sci. 1997;128(1):11–25.
Jamali KD, Ali SA. Yield and yield components with relation to plant height in semi-dwarf wheat. Pak J Bot. 2008;40(4):1805–8 http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.687.8119&rep=rep1&type=pdf.
Law CN, Snape JW, Worland AJ. The genetical relationship between height and yield in wheat. Heredity. 1978;40(1):133.
Denčić S, Kastori R, Kobiljski B, Duggan B. Evaluation of grain yield and its components in wheat cultivars and landraces under near optimal and drought conditions. Euphytica. 2000;113(1):43–52.
Knezevic D, Zecevic V, Stamenkovic S, Atanasijevic S, Milosevic B. Variability of number of kernels per spike in wheat cultivars (Triticum aestivum L.). J Cent Eur Agric. 2012;13(3):608–14 https://jcea.agr.hr/en/issues/article/1099.
Tuinstra M, Ejeta G, Goldsbrough P. Heterogeneous inbred family (HIF) analysis a method for develo** near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet. 1997;95:1005–11.
Zheng Z, Wang H, Chen G, Yan G, Liu C. A procedure allowing up to eight generations of wheat and nine generations of barley per annum. Euphytica. 2013;191(2):311–6.
Yan G, Liu H, Wang H, Lu Z, Wang Y, Mullan D, Hamblin J, Liu C. Accelerated generation of selfed pure line plants for gene identification and crop breeding. Front Plant Sci. 2017;8:1786. https://doi.org/10.3389/fpls.2017.01786.
Ma J, Yan GJ, Liu CJ. Development of near-isogenic lines for a major QTL on 3BL conferring Fusarium crown rot resistance in hexaploid wheat. Euphytica. 2011;183(2):147–52.
Wang X, Liu H, Mia MS, Siddique KHM, Yan G. Development of near-isogenic lines targeting a major QTL on 3AL for pre-harvest sprouting resistance in bread wheat. Crop Pasture Sci. 2018;69(9):864–72.
Mia MS, Liu H, Wang X, Yan G. Multiple near-isogenic lines targeting a QTL hotspot of drought tolerance showed contrasting performance under post-anthesis water stress. Front Plant Sci. 2019;10:271.
Habib A, Shabala S, Shabala L, Zhou M, Liu C. Near-isogenic lines developed for a major QTL on chromosome arm 4HL conferring Fusarium crown rot resistance in barley. Euphytica. 2015;209(3):555–63.
de la Torre F, Sampedro J, Zarra I, Revilla G. AtFXG1, an Arabidopsis gene encoding α-L-fucosidase active against fucosylated xyloglucan oligosaccharides. Plant Physiol. 2002;128(1):247–55.
Kurth EG, Peremyslov VV, Turner HL, Makarova KS, Iranzo J, Mekhedov SL, Koonin EV, Dolja VV. Myosin-driven transport network in plants. Proc Natl Acad Sci. 2017;114(8):E1385–94.
Van Damme M, Huibers RP, Elberse J, Van den Ackerveken G. Arabidopsis DMR6 encodes a putative 2OG-Fe (II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 2008;54(5):785–93.
Shorinola O, Balcárková B, Hyles J, Tibbits JFG, Hayden MJ, Holušova K, Valárik M, Distelfeld A, Torada A, Barrero JM, et al. Haplotype analysis of the pre-harvest sprouting resistance locus Phs-A1 reveals a causal role of TaMKK3-a in global germplasm. Front Plant Sci. 2017;8:1555.
Mares D, Mrva K, Cheong J, Williams K, Watson B, Storlie E, Sutherland M, Zou Y. A QTL located on chromosome 4A associated with dormancy in white- and red-grained wheats of diverse origin. Theor Appl Genet. 2005;111(7):1357–64.
Murray M, Thompson W. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res. 1980;8:4321–5.
Shorinola O, Bird N, Simmonds J, Berry S, Henriksson T, Jack P, Werner P, Gerjets T, Scholefield D, Balcarkova B, et al. The wheat Phs-A1 pre-harvest sprouting resistance locus delays the rate of seed dormancy loss and maps 0.3 cM distal to the PM19 genes in UK germplasm. J Exp Bot. 2016;67(14):4169–78.
Walker-Simmons M. Enhancement of ABA responsiveness in wheat embryos by high temperature. Plant Cell Environ. 1988;11:769–75.
Alaux M, Rogers J, Letellier T, Flores R, Alfama F, Pommier C, Mohellibi N, Durand S, Kimmel E, Michotey C. Linking the international wheat genome sequencing consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 2018;19(1):111.
Acknowledgements
The authors would like to thank Richard Wilkinson for his help with data collection of agronomic traits, and Professor Jacqueline Batley and Dr. Aneeta Pradhan for assistance in the 90 k SNP array genoty**.
Funding
The research was funded by Yipti Foundation Research Awards and Grants-in-Aid, and Global Innovation Linkage program (GIL53853) from Australian Department of Industry, Innovation and Science. The funding bodies were not involved in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
XW, HL, KHMS and GY conceived and designed the experiments; XW, GL and MSM conducted the experiments; XW, HL and MSM analysed the data; XW wrote the manuscript, and HL, GY and KHMS critically reviewed the manuscript. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, X., Liu, H., Liu, G. et al. Phenotypic and genotypic characterization of near-isogenic lines targeting a major 4BL QTL responsible for pre-harvest sprouting in wheat. BMC Plant Biol 19, 348 (2019). https://doi.org/10.1186/s12870-019-1961-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-019-1961-1