Abstract
Background
Chinese jujube (Ziziphus jujuba Mill.) is a non-climacteric fruit; however, the underlying mechanism of ripening and the role of abscisic acid involved in this process are not yet understood for this species.
Results
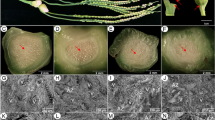
In the present study, a positive correlation between dynamic changes in endogenous ABA and the onset of jujube ripening was determined. Transcript analyses suggested that the expression balance among genes encoding nine-cis-epoxycarotenoid dioxygenase (ZjNCED3), ABA-8′-hydroxylase (ZjCYP707A2), and beta-glucosidase (ZjBG4, ZjBG5, ZjBG8, and ZjBG9) has an important role in maintaining ABA accumulation, while the expression of a receptor (ZjPYL8), protein phosphatase 2C (ZjPP2C4–8), and sucrose nonfermenting 1-related protein kinase 2 (ZjSnRK2–2 and ZjSnRK2–5) is important in regulating fruit sensitivity to ABA applications. In addition, white mature ‘Dongzao’ fruit were harvested and treated with 50 mg L− 1 ABA or 50 mg L− 1 nordihydroguaiaretic acid (NDGA) to explore the role of ABA in jujube fruit ripening. By comparative transcriptome analyses, 1103 and 505 genes were differentially expressed in response to ABA and NDGA applications on the 1st day after treatment, respectively. These DEGs were associated with photosynthesis, secondary, lipid, cell wall, and starch and sugar metabolic processes, suggesting the involvement of ABA in modulating jujube fruit ripening. Moreover, ABA also exhibited crosstalk with other phytohormones and transcription factors, indicating a regulatory network for jujube fruit ripening.
Conclusions
Our study further elucidated ABA-associated metabolic and regulatory processes. These findings are helpful for improving strategies for jujube fruit storage and for gaining insights into understand complex non-climacteric fruit ripening processes.
Similar content being viewed by others
Background
Chinese jujube (Ziziphus jujuba Mill.) is a popular fruit crop species that is native to China and is highly desired by consumers worldwide due to the abundant nutritional and health benefits of the fruit [1, 2]. However, the flesh jujube fruit has a very short shelf-life underlined by rapid dehydration or water-soaking deterioration within 2–3 days after harvest [3]. Therefore, fruit storage and quality maintenance have been among the most urgent challenges in the development of the jujube industry, whereas knowledge related to its ripening characterization and regulation is lacking. Over the past few decades, great strides have been made in elucidating the regulation of fruit ripening [4]; in particular, ethylene and abscisic acid (ABA) are recognized as the most important phytohormones that are directly or indirectly involved in the ripening of both climacteric and non-climacteric fruit [5, 6]. Recently, Chinese jujube has been characterized as a non-climacteric fruit, while a basal level of ethylene is still needed to maintain full fruit maturity [7]. These findings also reveal that the regulation of ripening is relatively complex and that there is a further need to explore these mechanisms to deepen our understanding of the ripening of Chinese jujube fruit.
With regard to ABA, the presence of dramatically increased levels in fruit during the onset of ripening has been reported in several fruit crop species, including grape [8], sweet cherry [9], cucumber [10], watermelon [11], and persimmon [5], which points to a role for ABA in triggering the onset of fruit ripening [8]. Moreover, applications of exogenous ABA and nordihydroguaiaretic acid (NDGA, an inhibitor of ABA biosynthesis) have enabled us to identify ABA-dependent pathways [12, 13]; increased numbers of research findings have suggested a positive role for ABA in promoting the metabolism and accumulation of soluble sugars [12, 14], formation of peel color [15, 16], and modification of cell wall catabolism [17], thereby accelerating ripening processes [5]. Fruit ripening is a highly integrated process that involves hormone control and crosstalk, as well as alterations to the numbers of transcripts of transcription factors (TFs) [61]. In the starch biosynthesis pathway, genes encoding ADP-glucose pyrophosphorylase and isoamylase were downregulated by ABA, while NDGA downregulated the expression of genes controlling starch degradation, including two alpha-amylase- and a beta-amylase-encoding genes. These results suggested that ABA was involved in starch metabolism, just as ZmEREB156 positively modulated starch biosynthesis via the synergistic effect of sucrose and ABA in maize [85]. For identification of differentially expressed genes (DEGs), an edgeR program [86] in OmicShare tools, a free online platform for data analysis (http://www.omicshare.com/tools), was used with a fold change (FC) threshold ≥2 and an false discovery rate (FDR) ≤ 0.05. The use of edgeR allowed comparative analysis within two replicates, and it had been used in several previous papers [12, 87, 88]. The functional enrichment of DEGs was determined using Gene Ontology (GO) and pathway analysis tools within the OmicShare platform [89]. We also used MapMan 3.6.0RC1 software to enrich the putative functional annotation of the DEGs [90, 91].
Quantitative real-time PCR validation for transcriptome expression levels
Total RNA was extracted using a plant RNA extraction kit (TaKaRa), and 200 ng of high-quality RNA was subsequently prepared for first-strand cDNA synthesis using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa). qPCR was then performed using a SYBR Premix Ex Taq II kit (TaKaRa) with a total volume of 10 μL, which comprised 1.0 μL of cDNA, 5.0 μL of SYBR premix solution, 0.4 μL of forward/reverse primers and 3.2 μL of dH2O. The thermal program for qPCR in a Roche LightCycler 96 system was set using the following conditions: 95 °C for 30 s; 40 cycles of amplification of 5 s at 95 °C, 30 s at 58 °C, and 30 s at 72 °C; and a default dissociation stage. The relative expression of each gene was normalized to that of a reference gene, ZjUBQ (Zhang et al. 2015), and was ultimately calculated using the 2-△Ct method (Livak and Schmittgen 2001). Sequences of the primers used for qPCR are listed in Additional file 10.
Statistical analysis
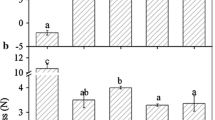
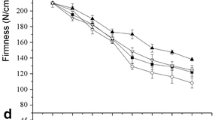
Statistical analysis was performed using the Duncan multiple range test (MRT) at the p < 0.05 level in SPSS 19.0. The error bars in the figures represent the standard deviations of three biological replicates.
Abbreviations
- AAO:
-
Abscisic aldehyde oxidase
- ABA:
-
Abscisic acid
- ABA2:
-
xanthoxin dehydrogenase
- ABA3:
-
molybdenum cofactor sulfurtransferase
- ACO:
-
1-aminocyclopropane-1-carboxylate oxidase
- ACS:
-
1-aminocyclopropane-1-carboxylate synthase
- AHP:
-
histidine-containing phosphotransferase
- AOG:
-
ABA-glucosyltransferase
- AP2/ERF:
-
APETALA2/ethylene-responsive element
- BG:
-
Beta-glucosidase
- BR:
-
Beginning red
- CYP707A:
-
ABA-8′-hydraxylase
- DAB:
-
Day after blooming
- DAT:
-
Day after treatment
- DEG:
-
Differentially expressed gene
- E:
-
Enlarging fruit
- EBF:
-
Ethylene insensitive 3-binding F-box
- EIN3/EIL:
-
Ethylene insensitive 3-like
- ERS:
-
Ethylene response
- ETR:
-
Ethylene receptor
- FC:
-
Fold change
- FDR:
-
False discovery rate
- FR:
-
Full-red
- GA:
-
Gibberenllin
- GA20ox:
-
GA-20-oxidase
- GA2ox:
-
GA-2-oxidase
- GH3:
-
Gretchen hagen 3
- GO:
-
Gene ontology
- HB:
-
Homeobox transcription factor
- HR:
-
Half-red
- HSF:
-
Heat-shock transcription factor
- KAO:
-
ent-kaurenoic acid hydroxylase
- LOG:
-
cytokinin riboside 5′-monophosphate phosphoribohydrolase
- MRT:
-
Multiple range test
- NCED:
-
Nine-cis-epoxycarotenoid dioxygenase
- NDGA:
-
Nordihydroguaiaretic acid
- PP2C:
-
Protein phosphatase 2C
- PYR/PYL/RCAR:
-
Pyrabatin resistance/pyrabatin resistance 1-like/regulatory component
- SnRK2:
-
Sucrose nonfermenting 1-related protein kinase 2
- TAA:
-
Tryptophan aminotransferase
- TF:
-
Transcription factor
- WM:
-
White mature
- YF:
-
Young fruit
- ZEP:
-
Zeaxanthin epoxidase
References
Gao QH, Wu CS, Wang M. The jujube (Ziziphus jujuba mill.) fruit. A review of current knowledge of fruit composition and health benefits. J Agric Food Chem. 2013;61(14):3351–63.
Yao S. Past, present, and futrue of jujubes- Chinese dates in the United States. Hortscience. 2013;48(6):672–80.
Wu H, Wang S, Zhu J, Meng X, Wang D. Postharvest treatments affecting storage quality of Chinese jujubes. In: Liu D, editor. Chinese dates: a traditional functional food. 1st ed. Boca Raton: CRC Press; 2016. p. 272–315.
Cherian S, Figueroa CR, Nair H. ‘Movers and shakers’ in the regulation of fruit ripening. A cross-dissection of climacteric versus non-climacteric fruit. J Exp Bot. 2014;65(17):4705–22.
Leng P, Yuan B, Guo Y. The role of abscisic acid in fruit ripening and responses to abiotic stress. J Exp Bot. 2014;65(16):4577–88.
Karlova R, Chapman N, David K, Angenent GC, Seymour GB, de Maagd RA. Transcriptional control of fleshy fruit development and ripening. J Exp Bot. 2014;65(16):4527–41.
Zhang Z, Huang J, Li X. Transcript analyses of ethylene pathway genes during ripening of Chinese jujube fruit. J Plant Physiol. 2018;224-225:1–10.
Sun LA, Zhang M, Ren J, Qi JX, Zhang GJ, Leng P. Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant Biol. 2010;10(1):257.
Luo H, ShengJie RD, Zhang C, Ding Y, Li Z, Sun Y, Ji K, Wang Y, Li Q, et al. The role of ABA in the maturation and postharvest life of a nonclimacteric sweet cherry fruit. Plant Growth Regul. 2014;33(2):373–83.
Wang Y, Wu Y, Duan C, Chen P, Li Q, Dai S, Sun L, Ji K, Sun Y, Xu W, et al. The expression profiling of the CsPYL, CsPP2C and CsSnRK2 gene families during fruit development and drought stress in cucumber. J Plant Physiol. 2012;169:1874-1882.
Wang Y, Guo S, Tian S, Zhang J, Ren Y, Sun H, Gong G, Zhang H, Xu Y. Abscisic acid pathway involved in the regulation of watermelon fruit ripening and quality trait evolution. PLoS One. 2017;12(6):e0179944.
Chen J, Mao L, Lu W, Ying T, Luo Z. Transcriptome profiling of postharvest strawberry fruit in response to exogenous auxin and abscisic acid. Planta. 2016;243(1):183–97.
Mou W, Li D, Luo Z, Mao L, Ying T. Transcriptomic analysis reveals possible influences of ABA on secondary metabolism of pigments, flavonoids and antioxidants in tomato fruit during ripening. PLoS One. 2015;10(6):e0129598.
Pilati S, Bagagli G, Sonego P, Moretto M, Brazzale D, Castorina G, Simoni L, Tonelli C, Guella G, Engelen K, et al. Abscisic acid is a major regulator of grape berry ripening onset: new insights into ABA signaling network. Front Plant Sci. 2017;8(1):1093.
Hu B, Li J, Wang D, Wang H, Qin Y, Hu G, Zhao J. Transcriptome profiling of Litchi chinensis pericarp in response to exogenous cytokinins and abscisic acid. Plant Growth Regul. 2017;84(3):437–50.
Li D, Li L, Luo Z, Mou W, Mao L, Ying T. Comparative transcriptome analysis reveals the influence of abscisic acid on the metabolism of pigments, ascorbic acid and folic acid during strawberry fruit ripening. PLoS One. 2015;10(6):e0130037.
Sun L, Sun YF, Zhang M, Wang L, Ren J, Cui MM, Wang YP, Ji K, Li P, Li Q, et al. Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol. 2012;158(1):283–98.
Wang Y, Tao X, Tang XM, **ao L, Sun JL, Yan XF, Li D, Deng HY, Ma XR. Comparative transcriptome analysis of tomato (Solanum lycopersicum) in response to exogenous abscisic acid. BMC Genomics. 2013;14(1):841.
Medina-Puche L, Blanco-Portales R, Molina-Hidalgo FJ, Cumplido-Laso G, Garcia-Caparros N, Moyano-Canete E, Caballero-Repullo JL, Munoz-Blanco J, Rodriguez-Franco A. Extensive transcriptomic studies on the roles played by abscisic acid and auxins in the development and ripening of strawberry fruits. Funct Integr Genomics. 2016;16(6):671–92.
Mou W, Li D, Bu J, Jiang Y, Khan ZU, Luo Z, Mao L, Ying T. Comprehensive analysis of ABA effects on ethylene biosynthesis and signaling during tomato fruit ripening. PLoS One. 2016;11(4):e0154072.
Murcia G, Pontin M, Piccoli P. Role of ABA and Gibberellin A3 on gene expression pattern of sugar transporters and invertases in Vitis vinifera cv. Malbec during berry ripening. Plant Growth Regul. 2017;84(2):275–83.
Estrada-Johnson E, Csukasi F, Pizarro CM, Vallarino JG, Kiryakova Y, Vioque A, Brumos J, Medina-Escobar N, Botella MA, Alonso JM, et al. Transcriptomic analysis in strawberry fruits reveals active auxin biosynthesis and signaling in the ripe receptacle. Front Plant Sci. 2017;8(1):889.
Wang Y, Wang Y, Ji K, Dai S, Hu Y, Sun L, Li Q, Chen P, Sun Y, Duan C, et al. The role of abscisic acid in regulating cucumber fruit development and ripening and its transcriptional regulation. Plant Physiol Biochem. 2013;64(5):70–9.
Kuhn N, Guan L, Dai ZW, Wu BH, Lauvergeat V, Gomes E, Li SH, Godoy F, Arce-Johnson P, Delrot S. Berry ripening: recently heard through the grapevine. J Exp Bot. 2014;65(16):4543–59.
Zhang M, Leng P, Zhang G, Li X. Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. J Plant Physiol. 2009;166(12):1241–52.
Zhao SL, Qi JX, Duan CR, Sun L, Sun YF, Wang YP, Ji K, Chen P, Dai SJ, Leng P. Expression analysis of the DkNCED1, DkNCED2 and DkCYP707A1 genes that regulate homeostasis of abscisic acid during the maturation of persimmon fruit. J Hortic Sci Biotechnol. 2012;87(2):165–71.
Ji K, Chen P, Sun L, Wang YP, Dai SJ, Li Q, Li P, Sun YF, Wu Y, Duan CR, et al. Non-climacteric ripening in strawberry fruit is linked to ABA, FaNCED2 and FaCYP707A1. Funct Plant Biol. 2012;39(4):351.
Ji K, Kai W, Zhao B, Sun Y, Yuan B, Dai S, Li Q, Chen P, Wang Y, Pei Y, et al. SlNCED1 and SlCYP707A2: key genes involved in ABA metabolism during tomato fruit ripening. J Exp Bot. 2014;65(18):5243–55.
Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011;157(1):188–99.
Nambara E, Marion-Poll A. Abscisic acid biosynthesis and metabolism. Annu Rev Plant Biol. 2005;56(1):156–85.
Rodrigo MJ, Alquezar B, Zacarias L. Cloning and characterization of two 9-cis-epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck). J Exp Bot. 2006;57(3):633–43.
Hou YL, Meng K, Han Y, Ban QY, Wang B, Suo JT, Lv JY, Rao JP. The persimmon 9-lipoxygenase gene DkLOX3 plays positive roles in both promoting senescence and enhancing tolerance to abiotic stress. Front Plant Sci. 2015;6(1):360–438.
Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M. Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004;134(4):1439–49.
Cai Y, Zhu P, Liu C, Zhao A, Yu J, Wang C, Li Z, Huang P, Yu M. Characterization and expression analysis of cDNAs encoding abscisic acid 8′-hydroxylase during mulberry fruit maturation and under stress conditions. Plant Cell, Tiss Org. 2016;127(1):237–49.
Xu ZY, Lee KH, Dong T, Jeong JC, ** JB, Kanno Y, Kim DH, Kim SY, Seo M, Bressan RA, et al. A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell. 2012;24(5):2184–99.
Li Q, Ji K, Sun Y, Luo H, Wang H, Leng P. The role of FaBG3 in fruit ripening and B. cinerea fungal infection of strawberry. Plant J. 2013;76(1):24–35.
Karppinen K, Hirvela E, Nevala T, Sipari N, Suokas M, Jaakola L. Changes in the abscisic acid levels and related gene expression during fruit development and ripening in bilberry (Vaccinium myrtillus L.). Phytochemistry. 2013;95(6):127–34.
Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH. Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol. 2007;143(2):745–58.
Gonzalez-Guzman M, Abia D, Salinas J, Serrano R, Rodriguez PL. Two new alleles of the abscisic aldehyde oxidase 3 gene reveal its role in abscisic acid biosynthesis in seeds. Plant Physiol. 2004;135(1):325–33.
Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T. Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 2004;45(11):1694–703.
**ong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13(9):2063–83.
Romero P, Lafuente MT, Rodrigo MJ. The Citrus ABA signalosome: identification and transcriptional regulation during sweet orange fruit ripening and leaf dehydration. J Exp Bot. 2012;63(13):4931–45.
Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q, Li P, Dai SJ, Leng P. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J Exp Bot. 2011;62(15):5659–69.
Chai YM, Jia HF, Li CL, Dong QH, Shen YY. FaPYR1 is involved in strawberry fruit ripening. J Exp Bot. 2011;62(14):5079–89.
Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25(3):295–303.
Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade a PP2Cs. Plant J. 2009;60(4):575–88.
Chen P, Sun YF, Kai WB, Liang B, Zhang YS, Zhai XW, Jiang L, Du YW, Leng P. Interactions of ABA signaling core components (SlPYLs, SlPP2Cs, and SlSnRK2s) in tomato (Solanum lycopersicon). J Plant Physiol. 2016;205(1):67–74.
Gambetta GA, Matthews MA, Shaghasi TH, McElrone AJ, Castellarin SD. Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta. 2010;232(1):219–34.
Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21(10):3170–84.
Li X. Mechanisms of the signal transduction for ethylene production in apple (Malus ×domestica) fruit development and ripening. Bei**g: China Agricultural University; 2014.
Han Y, Dang RH, Li JX, Jiang JZ, Zhang N, Jia MR, Wei LZ, Li ZQ, Li BB, Jia WS. SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2.6, an ortholog of OPEN STOMATA1, os a negative regulator of strawberry fruit development and ripening. Plant Physiol. 2015;167(3):915–30.
Kou X, He Y, Li Y, Chen X, Feng Y, Xue Z. Effect of abscisic acid (ABA) and chitosan/nano-silica/sodium alginate composite film on the color development and quality of postharvest Chinese winter jujube (Zizyphus jujuba mill. cv. Dongzao). Food Chem. 2019;270:385–94.
Creelman RA, Bell E, Mullet JE. Involvement of a lipoxygenase-like enzyme in abscisic acid biosynthesis. Plant Physiol. 1992;99(3):1258–60.
Wang X, Yin W, Wu J, Chai L, Yi H. Effects of exogenous abscisic acid on the expression of citrus fruit ripening-related genes and fruit ripening. Sci Hortic-Amsterdam. 2016;201(1):175–83.
Zhang M, Yuan B, Leng P. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot. 2009;60(6):1579–88.
Palumbo MC, Zenoni S, Fasoli M, Massonnet M, Farina L, Castiglione F, Pezzotti M, Paci P. Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. Plant Cell. 2014;26(12):4617–35.
Ramachandra Reddy A, Chaitanya KV, Vivekanandan M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J Plant Physiol. 2004;161(11):1189–202.
Peppi M, Fidelibus M, Dokoozlian N. Application timing and concentration of abscisic acid affect the quality of ‘Redglobe’ grapes. J Hort Sci B. 2007;82:304–10.
Koyama K, Sadamatsu K, Goto-Yamamoto N. Abscisic acid stimulated ripening and gene expression in berry skins of the cabernet sauvignon grape. Funct Integr Genomics. 2010;10(3):367–81.
Han X, Lu W, Wei X, Li L, Mao L, Zhao Y. Proteomics analysis to understand the ABA stimulation of wound suberization in kiwifruit. J Proteome. 2018;173(1):42–51.
Deytieux-Belleau C, Vallet A, Doneche B, Geny L. Pectin methylesterase and polygalacturonase in the develo** grape skin. Plant Physiol Biochem. 2008;46(7):638–46.
Huang HH, **e SD, **ao QL, Wei B, Zheng LJ, Wang YB, Cao Y, Zhang XG, Long TD, Hu YF et al. Sucrose and ABA regulate starch biosynthesis in maize through a novel transcription factor, ZmEREB156. Sci Rep 2016, 6:27590 (1):1.
Klee HJ, Giovannoni JJ. Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet. 2011;45(1):41–59.
Seymour GB, Ostergaard L, Chapman NH, Knapp S, Martin C. Fruit development and ripening. Annu Rev Plant Biol. 2013;64(1):219–41.
Guo HW, Ecker JR. Plant responses to ethylene gas are mediated by SCF (EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115(6):667–77.
Yang Y, Wu Y, Pirrello J, Regad F, Bouzayen M, Deng W, Li Z. Silencing Sl-EBF1 and Sl-EBF2 expression causes constitutive ethylene response phenotype, accelerated plant senescence, and fruit ripening in tomato. J Exp Bot. 2010;61(3):697–708.
McAtee P, Karim S, Schaffer R, David K. A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front Plant Sci. 2013;4(79):79.
Srivastava A, Handa AK. Hormonal regulation of tomato fruit development. A molecular perspective. J Plant Growth Regul. 2005;24(2):67–82.
Symons GM, Chua YJ, Ross JJ, Quittenden LJ, Davies NW, Reid JB. Hormonal changes during non-climacteric ripening in strawberry. J Exp Bot. 2012;63(13):4741–50.
Kappel F, MacDonald RA. Gibberellic acid increases fruit firmness, fruit size, and delays maturity of 'sweetheart' sweet cherry. J Am Pomol Soc. 2002;56(4):210–22.
Wu JX, ll F, Yi HL. Genome-wide identification of the transcription factors involved in Citrus fruit ripening from the transcriptomes of a late-ripening sweet orange mutant and its wild type. PLoS One. 2016;11(4):e0154330.
Zhang Z, Li XG. Genome-wide identification of AP2/ERF superfamily genes and their expression during fruit ripening of Chinese jujube. Sci Rep-Uk. 2018(8):15162.
Nicolas P, Lecourieux D, Kappel C, Cluzet S, Cramer G, Delrot S, Lecourieux F. The basic leucine zipper transcription factor ABSCISIC ACID RESPONSE ELEMENT-BINDING FACTOR2 is an important transcriptional regulator of abscisic acid-dependent grape berry ipening processes. Plant Physiol. 2014;164(1):365–83.
**e XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012;35(11):1884–97.
Zhang H, Liu YP, Wen F, Yao DM, Wang L, Guo J, Ni L, Zhang A, Tan MP, Jiang MY. A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J Exp Bot. 2014;65(20):5795–809.
Ma N, Feng H, Meng X, Li D, Yang D, Wu C, Meng Q. Overexpression of tomato SlNAC1transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 2014;14(1):351.
Shi WG, Li H, Liu TX, Polle A, Peng CH, Luo ZB. Exogenous abscisic acid alleviates zinc uptake and accumulation in Populus x canescens exposed to excess zinc. Plant Cell Environ. 2015;38(1):207–23.
Pan X, Welti R, Wang X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc. 2010;5(6):986–92.
Fugate KK, Suttle JC, Campbell LG. Ethylene production and ethylene effects on respiration rate of postharvest sugarbeet roots. Postharvest Biol Tec. 2010;56(1):71–6.
Huang J, Zhang CM, Zhao X, Fei ZJ, Wan KK, Zhang Z, Pang XM, Yin X, Bai Y, Sun XQ, et al. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet. 2016;12(12):e1006433.
Letunic I, Doerks T, Bork P. SMART. Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(Database issue):D257–60.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4.
Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11.
Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Map** and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40.
Li X, An M, **a Z, Bai X, Wu Y. Transcriptome analysis of watermelon (Citrullus lanatus) fruits in response to Cucumber green mottle mosaic virus (CGMMV) infection. Sci Rep-UK. 2017;7(1):16747.
Zhai R, Feng Y, Wang H, Zhan X, Shen X, Wu W, Zhang Y, Chen D, Dai G, Yang Z, et al. Transcriptome analysis of rice root heterosis by RNA-Seq. BMC Genomics. 2013;14:19.
Li S, Zhu S, Jia Q, Yuan D, Ren C, Li K, Liu S, Cui Y, Zhao H, Cao Y, et al. The genomic and functional landscapes of developmental plasticity in the American cockroach. Nat Commun. 2018;9(1):1008.
Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37(6):914–39.
Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, maize. Plant Cell Environ. 2009;32(9):1211–29.
Acknowledgements
Not applicable.
Funding
This work was financially supported by the National Key R&D Program of China (2018YFD1000607), Ministry of Science and Technology of the People’s Republic of China. The supporters did not play any role in the design, collection, analysis, interpretation of the relevant data, or in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additional files.
Author information
Authors and Affiliations
Contributions
ZZ and XL conceived and designed the experiments. ZZ performed samples preparation and conducted all the experiments, data analyses and wrote the manuscript. CK and SZ participated in determination of fruit respiration and ethylene production during postharvest storage. ZZ and XL contributed to the discussion. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Statistic information of RNA-seq data. (XLSX 11 kb)
Additional file 2:
Statistic information of map** data. (XLSX 11 kb)
Additional file 3:
GO enrichment for DEGs at DAT1. (XLSX 17 kb)
Additional file 4:
KEGG pathway enrichment for DEGs (Level3) at DAT1. (XLSX 27 kb)
Additional file 5:
DEGs related to metabolism pathways by MapMan. (XLSX 68 kb)
Additional file 6:
DEGs related to hormone metabolism and signaling. (XLSX 21 kb)
Additional file 7:
DEGs related to transcription factors involved in ripening regulation. (XLSX 27 kb)
Additional file 8:
RT-qPCR validation of digital expression patterns revealed by RNA sequencing. A number of 17 genes were selected to validate the transcriptomic expressions by qPCR. The correlation coefficient between the RNA-seq data and relative expression ranged from 0.838–1.0, thereby confirming the reliability of the RNA data. (DOCX 241 kb)
Additional file 9:
Phylogenetic analyses for NCED, CYP707A, BG, PYR/PYL/RCAR, PP2C, and SnRK2 genes. The trees were generated by the multiple alignments with putative proteins from Arabidopsis, grape, and tomato which were uploaded in the KEGG database using MEGA 7.0. The Bootstrap value was set into 1000 (Kumar et al. 2016). (DOCX 983 kb)
Additional file 10:
Sequences for ABA pathway genes. (XLSX 124 kb)
Additional file 11:
Accession of clean reads submitted to sequence read archives (SRA) of NCBI. (XLSX 10 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, Z., Kang, C., Zhang, S. et al. Transcript analyses reveal a comprehensive role of abscisic acid in modulating fruit ripening in Chinese jujube. BMC Plant Biol 19, 189 (2019). https://doi.org/10.1186/s12870-019-1802-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-019-1802-2