Abstract
Background
The leaves of tea plants (Camellia sinensis) are used to produce tea, which is one of the most popular beverages consumed worldwide. The nutritional value and health benefits of tea are mainly related to three abundant characteristic metabolites; catechins, theanine and caffeine. Weighted gene co-expression network analysis (WGCNA) is a powerful system for investigating correlations between genes, identifying modules among highly correlated genes, and relating modules to phenotypic traits based on gene expression profiling. Currently, relatively little is known about the regulatory mechanisms and correlations between these three secondary metabolic pathways at the omics level in tea.
Results
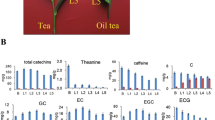
In this study, levels of the three secondary metabolites in ten different tissues of tea plants were determined, 87,319 high-quality unigenes were assembled, and 55,607 differentially expressed genes (DEGs) were identified by pairwise comparison. The resultant co-expression network included 35 co-expression modules, of which 20 modules were significantly associated with the biosynthesis of catechins, theanine and caffeine. Furthermore, we identified several hub genes related to these three metabolic pathways, and analysed their regulatory relationships using RNA-Seq data. The results showed that these hub genes are regulated by genes involved in all three metabolic pathways, and they regulate the biosynthesis of all three metabolites. It is notable that light was identified as an important regulator for the biosynthesis of catechins.
Conclusion
Our integrated omics-level WGCNA analysis provides novel insights into the potential regulatory mechanisms of catechins, theanine and caffeine metabolism, and the identified hub genes provide an important reference for further research on the molecular biology of tea plants.
Similar content being viewed by others
Background
Tea, produced from the leaves of the tea plant, Camellia sinensis (L.), belonging to family Theaceae, is one of the most popular natural non-alcoholic beverages consumed worldwide. To date, nearly 4000 bioactive compounds have been identified in tea [1] including catechins, caffeine, theanine and volatile oils [2]. Catechins generally contain six different monomers, namely catechin (C), gallocatechin (GC), epicatechin (EC), epigallocatechin (EGC), epicatechin gallate (ECG) and epigallocatechin gallate (EGCG) [3]. Catechins, caffeine and theanine are the main three characteristic biologically active compounds in tea [4]. They are not only important contributors to flavour, but also have beneficial effects on human health due to their autoxidation and anticancer activity [5] and their ability to lower blood pressure [6], prevent cardiovascular diseases [7], and assist weight loss [8].
Gene co-expression network analysis (GCNA) is a genetic approach for analysing correlations between genes using large-scale gene expression profiling data that is especially useful for investigating relationships between functional modules and phenotypic traits [9, 10]. Weighted GCNA (WGCNA) is one of the most popular GCNA-based approaches, and this correlation-based technique describes and visualises co-expression networks between genes using transcriptomic data [11]. This technique has been successfully utilized to identify the gene modules in Arabidopsis and rice that are related to drought and bacterial stress [12]. Module assignment in WGCNA is a flexible process which reduces the complexity of a dataset from hundreds of genes to a smaller number of modules.
Researchers have focused on the molecular mechanisms involved in plant growth, development [13, 14] and the production of secondary metabolites [15] in tea plants. Regulatory mechanisms underlying secondary metabolite biosynthesis, particularly those related to catechins, theanine and caffeine, have been explored at the molecular level. Recent advances in next-generation sequencing of RNA [16] have been accompanied by an increase in the amount of available transcriptomic data from different tissues of tea plants [17], from different species of the genus Camellia [18], and from plants grown under different stress conditions [55], which helped to elucidate the complex biological functions of genes. Based on the BLAST results from the NR database, GO annotation was carried out using the Blast 2 GO program (version 2.3.4) [56].
Identification of gene expression and DEGs
Expression levels of unigenes were calculated using the FPKM method. Firstly, reads were mapped to unigene datasets by Bowtie2 (version 2.1.0, http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) at a sensitive setting. Based on the Bowtie results, FPKM values for each unigene were subsequently calculated by RESM (version 1.2.29) [57] with default parameters. DEGs were identified based on the method described by Audic et al. [58]. Genes with|log2ratio| ≥ 1 and false discovery rate (FDR) < 0.05 were identified as DEGs.
Construction of gene co-expression networks
Gene co-expression networks were constructed using the WGCNA approach with R packages (version 3.2.2). DEGs expressed in at least one pairwise comparison in ten tissues were retained for co-expression network construction by WGCNA analysis [11]. All tissues were initially clustered to analyse the sample height. Following application of the scale-free topology criterion described previously, a soft threshold of 30 was chosen. Based on the topological overlap-based dissimilarity measure [59], unigenes were first hierarchically clustered, and the gene dendrogram was used for module detection by the dynamic tree cut method (mergeCutHeight = 0.25, minModuleSize = 30). In the weighted gene co-expression network, gene connectivity was based on the edge weight (ranging from 0 to 1) determined by the topology overlap measure, which reflects the strength of the communication between two genes. The weights across all edges of a node were summed and used to define the level of connectivity, and nodes with high connectivity were considered hub genes.
Identification of content-related modules
To identify modules associated with catechins, theanine and caffeine, we first calculated the module eigengenes of each module, then correlated these with the catechin, theanine and caffeine content using Pearson’s correlation coefficients and an asymptotic confidence interval based on Fisher’s Z transformation. Modules with p-values < 0.05 were identified as content-related modules. To further characterise these modules, enrichment of annotated unigenes in each content-related module was investigated using the phyper function within the R platform based on KEGG pathway annotation, and q-value or FDR corrections were applied by multiple testing [60]. We defined KEGG pathways with a q-value or FDR < 0.05 as significantly enriched [61].
Module hub gene selection and visualisation
The most central and connected genes, involved in numerous interactions, were considered hub genes [62], which are likely to play a more important role in a given module than other genes in the overall co-expression network. In this study, we categorised the top 2% of the most highly connected genes in a module as hub genes based on the size of the module. Co-expression interactions and patterns of hub genes were visualised using Cytoscape [63].
qPCR validation of selected unigenes
In order to evaluate the assembly quality of RNA-seq data, the expression patterns of eight selected transcripts were monitored by qPCR. RNA samples were isolated from samples using the CTAB method [45], and total RNA was reverse-transcribed into single-stranded cDNAs using a reverse transcription kit for real-time PCR (TaKaRa). Detailed information (unigene IDs and primer sequences) related to the selected transcripts used for qPCR is listed in Additional file 5. PCR amplification was performed according to the manufacturer’s instructions using a CFX96TM real-time PCR system (Bio-Rad) with an annealing temperature of 60 °C. The housekee** gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference gene, and relative expression levels of target genes were calculated using the 2ΔCt method [64]. All qPCRs were analysed using three technical and three biological replicates.
Abbreviations
- AGE:
-
Agarose gel electrophoresis
- C:
-
Catechin
- COG:
-
Clusters of Orthologous Groups of Proteins
- CS_FR:
-
Fruit
CS-R
Root
- CS-B1:
-
Apical bud
- CS-B2:
-
Apical bud
- CS-FL:
-
Flower
- CS-S:
-
Stem
- CS-SL:
-
Mature leaf in summer
- CS-WL:
-
Mature leaf in winter
- CS-YL1:
-
First young leaf
- CS-YL2:
-
Second young leaf
- EC:
-
Epicatechin
- ECG:
-
Epicatechin gallate
- EGC:
-
epigallocatechin
- EGCG:
-
Epigallocatechin gallate
- F3’5’H:
-
Flavonoid 3’,5’-hydroxylase
- FLS:
-
Flavonol synthase
- FPKM:
-
Fragment Per Kilobase of exon model per Million mapped reads
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- GC:
-
Gallocatechin
- GO:
-
Gene Ontology
- GS:
-
Glutamine synthetase
- HCT:
-
Shikimate O-hydroxycinnamoyl transferase
- HPLC:
-
High-performance liquid chromatography
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- Nr:
-
Non-redundant protein database
- Nt:
-
Non-redundant nucleotide database
- qPCR:
-
Quantitative real-time polymerase chain reaction
- Swiss-Prot:
-
Annotated protein sequence database
- WGCNA:
-
Weighted gene co-expression network analysis
- βG:
-
Beta-glucosidase
References
Mahmood T, Akhtar N, Khan BA. The morphology, characteristics, and medicinal properties of Camellia sinensis' tea. J Med Plant Res. 2010;4(19):2028–33.
Mamati GE, Liang Y, Lu J. Expression of basic genes involved in tea polyphenol synthesis in relation to accumulation of catechins and total tea polyphenols. J Sci Food Agric. 2006;86(3):459–64.
Punyasiri P, Abeysinghe I, Kumar V, Treutter D, Duy D, Gosch C, Martens S, Forkmann G, Fischer T. Flavonoid biosynthesis in the tea plant Camellia sinensis: properties of enzymes of the prominent epicatechin and catechin pathways. Arch Biochem Biophys. 2004;431(1):22–30.
Liang YR, Ma WY, Lu JL, Wu Y. Comparison of chemical compositions of Ilex latifolia Thumb and Camellia sinensis L. Food Chem. 2001;75(3):339–43.
Sasazuki S, Tamakoshi A, Matsuo K, Ito H, Wakai K, Nagata C, Mizoue T, Tanaka K, Tsuji I, Inoue M. Green tea consumption and gastric cancer risk: an evaluation based on a systematic review of epidemiologic evidence among the Japanese population. Jpn J Clin Oncol. 2012;42(4):335.
Hodgson JM, Croft KD, Woodman RJ, Puddey IB, Fuchs D, Draijer R, Lukoshkova E, Head GA. Black tea lowers the rate of blood pressure variation: a randomized controlled trial. Am J Clin Nutr. 2013;97(5):943–50.
Hollman PC, Feskens EJ, Katan MB. Tea flavonols in cardiovascular disease and cancer epidemiology. Proc Soc Exp Biol Med. 1999;220(4):198–202.
Auvichayapat P, Prapochanung M, Tunkamnerdthai O, Sripanidkulchai BO, Auvichayapat N, Thinkhamrop B, Kunhasura S, Wongpratoom S, Sinawat S, Hongprapas P. Effectiveness of green tea on weight reduction in obese Thais: a randomized, controlled trial. Physiol Behav. 2008;93(3):486–91.
Fuller T, Langfelder P, Presson A, Horvath S. Review of weighted gene coexpression network analysis. In: Handbook of Statistical Bioinformatics: Springer; 2011. p. 369–88.
Ruan J, Dean AK, Zhang W. A general co-expression network-based approach to gene expression analysis: comparison and applications. BMC Syst Biol. 2010;4(1):8.
Steve H, Peter L. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9(1):559.
Shaik R, Ramakrishna W. Genes and co-expression modules common to drought and bacterial stress responses in Arabidopsis and rice. PLoS One. 2013;8(10):e77261.
Obanda M, Owuor PO. Impact of shoot maturity on chlorophyll content, composition of volatile flavour compounds and plain black tea chemical quality parameters of clonal leaf. J Sci Food Agric. 1995;69(4):529–34.
Pokinda O, Martin O, Hastingse N, Wilsonl M. Influence of region of production on clonal black tea chemical characteristics. Food Chem. 2008;108(1):263–71.
Nagar PK, Sood S. Changes in endogenous auxins during winter dormancy in tea (Camellia sinensis L.) O. Kuntze. Acta Physiol Plant. 2006;28(2):165–9.
Wang Z, Gerstein M, Snyder M, Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63.
Shi CY, Hua Y, Wei CL, Yu O, Zhang ZZ, Jiang CJ, Sun J, Li YY, Qi C, Tao X. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics. 2011;12(1):131.
Tai Y, Wei C, Yang H, Zhang L, Chen Q, Deng W, Wei S, Zhang J, Fang C, Ho C. Transcriptomic and phytochemical analysis of the biosynthesis of characteristic constituents in tea (Camellia sinensis) compared with oil tea (Camellia oleifera). BMC Plant Biol. 2015;15(1):190.
**n-Chao Wang Q-YZ, Ma C-l, Zhang Z-H, Cao H-l, Kong Y-M, Yue C, Hao X-Y, Chen L, Ma J-Q, ** J-Q, li X, Yang Y-J. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics. 2013;14(1):1–15.
Thirugnanasambantham K, Prabu G, Palanisamy S, Chandrabose SR, Mandal AK. Analysis of dormant bud (Banjhi) specific transcriptome of tea (Camellia sinensis (L.) O. Kuntze) from cDNA library revealed dormancy-related genes. Appl Biochem Biotechnol. 2013;169(4):1405–17.
Li CF, Zhu Y, Yu Y, Zhao QY, Wang SJ, Wang XC, Yao MZ, Luo D, Li X, Chen L. Global transcriptome and gene regulation network for secondary metabolite biosynthesis of tea plant (Camellia sinensis). BMC Genomics. 2015;16(1):560.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52.
Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, Parvizi B. TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19(5):651.
Rau A, Gallopin M, Celeux G, Jaffrézic F. Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics. 2013;29(17):2146–52.
Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357.
Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5(12):e1000598.
Bai S, Sun Y, Qian M, Yang F, Ni J, Tao R, Lin L, Shu Q, Dong Z, Teng Y. Transcriptome analysis of bagging-treated red Chinese sand pear peels reveals light-responsive pathway functions in anthocyanin accumulation. Sci Rep. 2017;7(1):63.
Wu S, Lei J, Chen G, Chen H, Cao B, Chen C. De novo Transcriptome assembly of Chinese kale and global expression analysis of genes involved in Glucosinolate metabolism in multiple tissues. Front Plant Sci. 2017;8:92.
Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271.
Feldheim W, Yongvanit P, Cummings PH. Investigation of the presence and significance of theanine in the tea plant. J Sci Food Agric. 1986;37(6):527–34.
Tanaka T, Watarumi S, Fujieda M, Kouno I. New black tea polyphenol having N-ethyl-2-pyrrolidinone moiety derived from tea amino acid theanine: isolation, characterization and partial synthesis. Food Chem. 2005;93(1):81–7.
Emmersen J. Generating unigene collections of expressed sequence tag sequences for use in mass spectrometry identification. Methods Mole Biol (Clifton, NJ). 2007;367:77.
Stasolla C, Katahira R, Thorpe TA, Ashihara H. Purine and pyrimidine nucleotide metabolism in higher plants. J Plant Physiol. 2003;160(11):1271–95.
Loïc L, Debeaujon I, Routaboul J-M, Baudry A, Pourcel L, Nesi N, Caboche M. Genetics and biochemistry of seed flavonoids. Plant Biol. 2006;57(57):405–30.
Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20(3):176–85.
Shimada Y, Nakano-Shimada R, Ohbayashi M, Okinaka Y, Kiyokawa S, Kikuchi Y. Expression of chimeric P450 genes encoding flavonoid-3’,5’-hydroxylase in transgenic tobacco and petunia plants 1. FEBS Lett. 1999;461(3):241–5.
Liu Y, Gao L, Liu L, Yang Q, Lu Z, Nie Z, Wang Y, **a T. Purification and characterization of a novel Galloyltransferase involved in Catechin Galloylation in the tea plant (Camellia sinensis). J Biol Chem. 2012;287(53):44406–17.
Zhang WJ, Liang YR, Zhang FZ, Chen CS, Zhang YG, Chen RB, Weng BQ. Effects on the yield and quality of oolong tea by covering with shading net. J Tea Sci. 2004;24(4):276–82.
Kangmo K, Jungnam C, Jiyoung K, Jeongkee K, Langgook Y, Sangjun L, Hong YS, Choonghwan L. Metabolomics analysis reveals the compositional differences of shade grown tea (Camellia sinensis L.). J Agric Food Chem. 2009;58(1):418–26.
Wang YS, Gao LP, Shan Y, Liu YJ, Tian YW, **a T. Influence of shade on flavonoid biosynthesis in tea (Camellia sinensis (L.) O. Kuntze). Sci Hortic. 2012;141(3):7–16.
Gedda L. Light-induced expression of genes involved in phenylpropanoid biosynthetic pathways in callus of tea (Camellia sinensis (L.) O. Kuntze). Sci Hortic. 2012;133(1):72–83.
Tai Y, Wang H, Wei C, Su L, Li M, Wang L, Dai Z, Shao L, Jiang J, Luo M. Construction and characterization of a bacterial artificial chromosome library for Camellia sinensis. Tree Genet Genomes. 2017;13(4):89.
Wang L, Gong LH, Chen CJ, Han HB, Li HH. Column-chromatographic extraction and separation of polyphenols, caffeine and theanine from green tea. J Crohns Colitis. 2016;10(8):873.
Yin-Hua LI, Liu ZH, Huang JA, Gong YS. Determination of Theanine in tea by reversed-phase high performance liquid chromatography-evaporative light-scattering detector. J Tea Sci. 2005;25(3):225–8.
Shi CY, Wan XC, Jiang CJ, Sun J. Method for high-quality total RNA isolation from tea plants (Camellia sinensis (L.) O. Kuntze). J Anhui Agric Univ. 2007;34(3):360–3.
Liu S, Shi XX, Jiang YD, Zhu ZJ, Qian P, Zhang MJ, Yu H, Zhu QZ, Gong ZJ, Zhu ZR. De novo analysis of the Tenebrio molitor (Coleoptera: Tenebrionidae) transcriptome and identification of putative glutathione S -transferase genes. Appl Entomol Zool. 2014;50(1):1–9.
Gu XC, Zhang YN, Kang K, Dong SL, Zhang LW. Antennal Transcriptome Analysis of Odorant Reception Genes in the Red Turpentine Beetle (RTB), Dendroctonus valens. PLoS One. 2015;10(5):e0125159.
Tsanakas GF, Manioudaki ME, Economou AS, Kalaitzis P. De novo transcriptome analysis of petal senescence in Gardenia jasminoides Ellis. BMC Genomics. 2014;15(1):554.
Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z. SOAPnuke: a MapReduce acceleration supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2017;7(1):1–6.
Shao Y, Wei J, Wu F, Zhang H, Yang D, Liang Z, ** W. DsTRD: Danshen transcriptional resource database. PLoS One. 2016;11(2):e0149747.
Li S, Liu H, Bai J, Zhu X. Transcriptome assembly and identification of genes and SNPs associated with growth traits in largemouth bass (Micropterus salmoides). Genetica. 2017;145(2):175–87.
Łabaj PP, Leparc GG, Linggi BE, Markillie LM, Wiley HS, Kreil DP. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Bioinformatics. 2011;27(13):i383–91.
Sam LT, Lipson D, Raz T, Cao X, Thompson J, Milos PM, Robinson D, Chinnaiyan AM, Kumar-Sinha C, Maher CA. A comparison of single molecule and amplification based sequencing of cancer transcriptomes. PLoS One. 2011;6(3):e17305.
Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Conesa A, Götz S, García-Gómez JM, Terol J, Manuel N, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. Bmc Bioinform. 2011;12(1):323.
Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–95.
Zhang B, Horvath S. A General Framework For Weighted Gene Co-Expression Network Analysis. Stat Appl Genet Mol Biol. 2005;4(1) Article17
Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–8.
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):277–80.
Liu J, Ling J, Tu X. Weighted gene co-expression network analysis identifies specific modules and hub genes related to coronary artery disease. BMC Cardiovasc Disord. 2016;16(1):1–8.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.
Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, Mannucci S, De Nisi MC, Toscano M, Malagnino V. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol. 2007;30(2):429–36.
Acknowledgements
We would like to thank the native English speaking scientists of Elixigen Company (Huntington Beach, California) for editing our manuscript.
Funding
This work was supported by the Special Innovative Province Construction in Anhui Province [grant number 15czs08032], the Special Project for Central Guiding Science and Technology Innovation of Region in Anhui Province [grant number 2016080503B024], and the Natural Science Foundation of Anhui Province [grant number 31170283], and the Youth Foundation of Anhui Agricultural University [grant number 2016ZR012]. The design of this study and the manuscript writing were supported by the Science and Technology Project of Anhui Province, China [grant number 13Z03012]. And the collection, analysis, and interpretation of data used in this study were supported by youth Foundation of Anhui Agricultural University [grant number 2016ZR012].
Availability of data and materials
The Illumina RNA-seq data generated from C. sinensis are available from the NCBI SRA database (http://trace.ncbi.nlm.nih.gov/Traces/sra) under accessions SRR1928149 and SRP056466.
Author information
Authors and Affiliations
Contributions
XCW and CLW participated in the research design. YLT and CL analysed data and drafted the manuscript, SWY and JMS performed most of the experiments, and CXG, YY, BH and ZYL provided technical assistance. HY participated in material collection and manuscript revision. EHX contributed to revising the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Six-year-old tea plants (C. sinensis cv. Shuchazao) were used in this study. The field experiment was performed in a typical tea-producing garden at De Chang Fabrication Base in Anhui Province, China (Shucheng, latitude 31.3 N, longitude 117.2E) under natural conditions. Collection of plant materials complied with the institutional, national and international guidelines. No specific permits were required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Clustering dendrogram of samples based on gene expression. (PDF 99 kb)
Additional file 2:
Modules significantly (p < 0.05) correlated with characteristic components in tea. (XLSX 11 kb)
Additional file 3:
Functional analysis of unigenes in blue, red and magenta modules enriched in the photosynthesis pathway. (XLSX 12 kb)
Additional file 4:
Hub genes for each highly significant content-related module. (XLSX 54 kb)
Additional file 5:
Unigene IDs and primer sequences for selected transcripts used for qPCR validation. (XLSX 9 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tai, Y., Liu, C., Yu, S. et al. Gene co-expression network analysis reveals coordinated regulation of three characteristic secondary biosynthetic pathways in tea plant (Camellia sinensis). BMC Genomics 19, 616 (2018). https://doi.org/10.1186/s12864-018-4999-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-018-4999-9